ggalign() is similar to ggplot in that it initializes a ggplot data and
mapping. ggalign() allowing you to provide data in various formats,
including matrices, data frames, or simple vectors. By default, it will
inherit from the layout. If a function, it will apply with the layout matrix.
ggalign() focuses on integrating plots into a layout by aligning the axes.
Arguments
- data
The following options can be used:
NULL: No data is set.waiver(): Inherits the data from the layout matrix.A
function(including purrr-like lambda syntax): Applied to the layout matrix to transform the data before use. To transform the final plot data, please usescheme_data().A
matrix,data.frame, or atomic vector.
- mapping
Default list of aesthetic mappings to use for plot. If not specified, must be supplied in each layer added to the plot.
- ...
<dyn-dots> Additional arguments passed to
fortify_data_frame().- size
The relative size of the plot, can be specified as a
unit(). Note that forcircle_layout(), all size values will be interpreted as relative sizes, as this layout type adjusts based on the available space in the circular arrangement.- no_axes
Logical; if
TRUE, removes axes elements for the alignment axis usingtheme_no_axes(). By default, will use the option-"ggalign.align_no_axes".- active
A
active()object that defines the context settings when added to a layout.
ggplot2 specification
ggalign initializes a ggplot object. The underlying data is created using
fortify_data_frame(). Please refer to it for more details.
When aligning discrete variables, ggalign() always applies a default
mapping for the axis of the data index in the layout. Specifically:
aes(y = .data$.y)is used for the horizontalstack_layout()(including left and right annotations).aes(x = .data$.x)is used for the verticalstack_layout()(including top and bottom annotations) andcircle_layout().
The following columns will be added to the data frame to align discrete variables:
.panel: The panel for the aligned axis. Refers to thex-axisfor verticalstack_layout()(including top and bottom annotations), and they-axisfor horizontalstack_layout()(including left and right annotations)..names(vec_names()) and.index(vec_size()/NROW()): Character names (if available) and the integer index of the original data..x/.yand.discrete_x/.discrete_y: Integer indices forx/ycoordinates, and a factor of the data labels (only applicable when names exist).
It is recommended to use .x/.y, or .discrete_x/.discrete_y as the
x/y mapping.
If the data inherits from quad_layout()/ggheatmap(), additional
columns will be added:
.extra_panel: Provides the panel information for the column (left or right annotation) or row (top or bottom annotation)..extra_index: The index information for the column (left or right annotation) or row (top or bottom annotation).
Discrete Axis Alignment
It is important to note that we consider rows as observations, meaning
vec_size(data)/NROW(data) must match the number of observations along the
axis used for alignment (x-axis for a vertical stack layout, y-axis for a
horizontal stack layout).
Examples
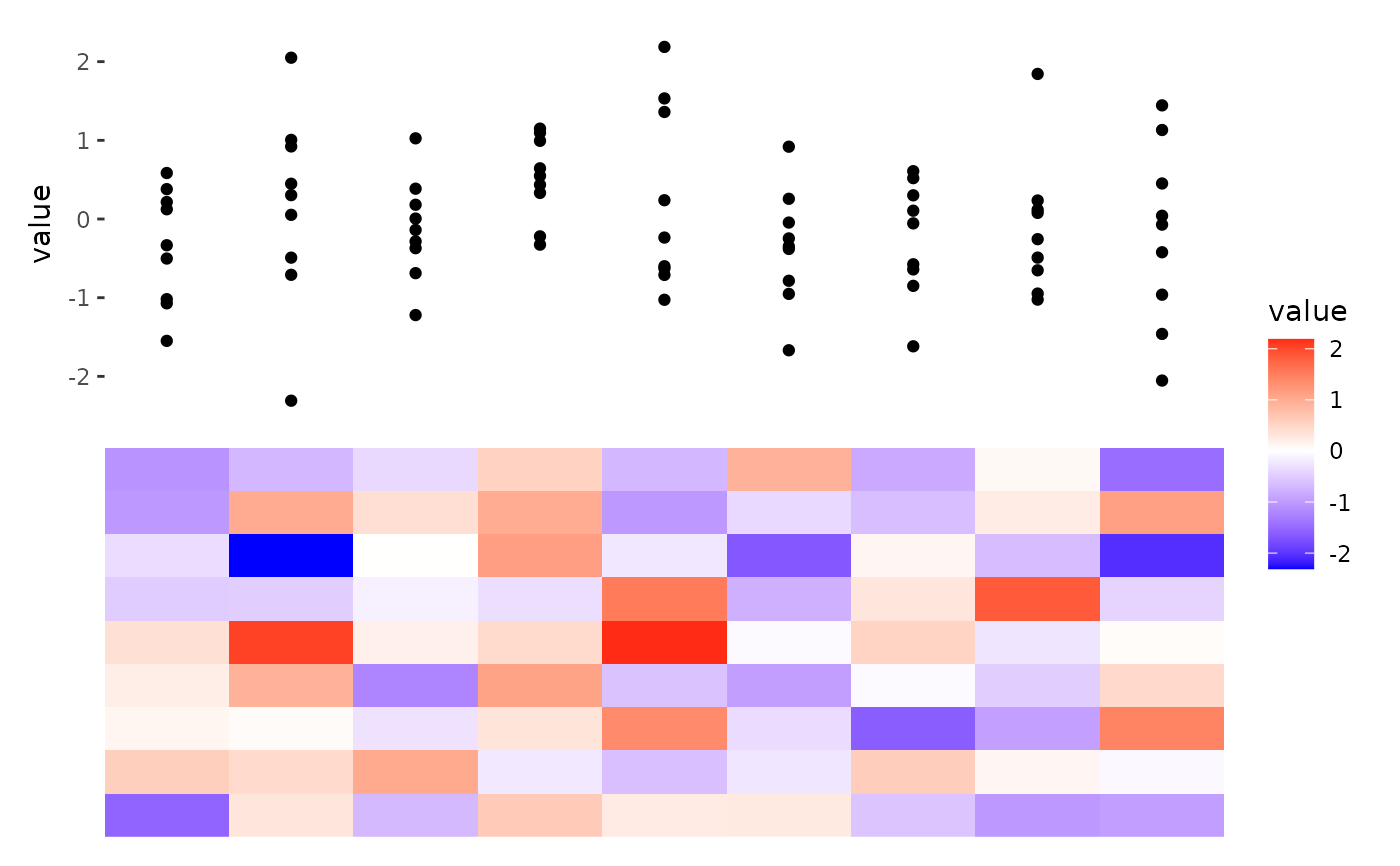
ggheatmap(matrix(rnorm(81), nrow = 9)) +
anno_top() +
ggalign() +
geom_point(aes(y = value))
#> → heatmap built with `geom_tile()`
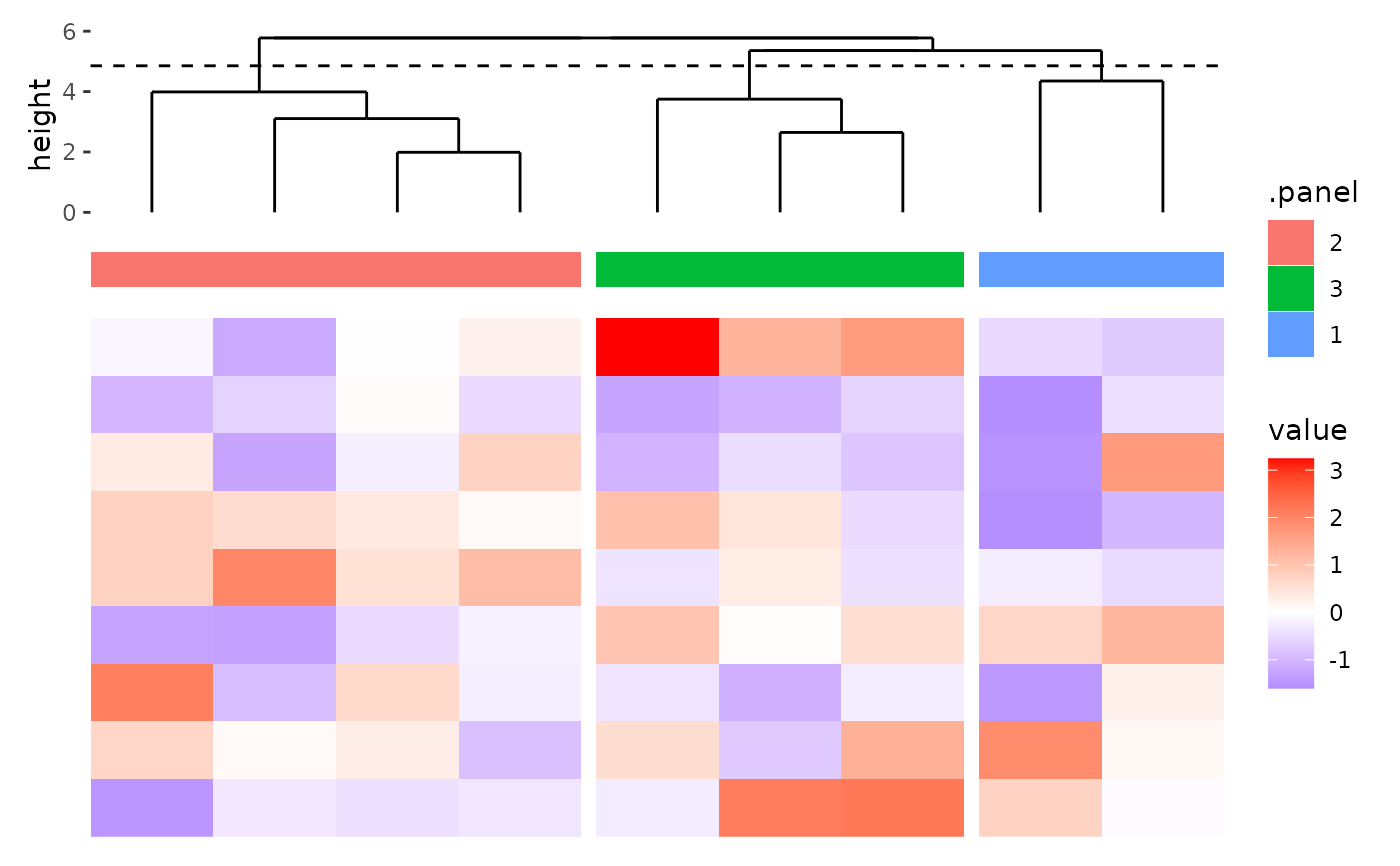
 ggheatmap(matrix(rnorm(81), nrow = 9)) +
anno_top(size = 0.5) +
align_dendro(k = 3L) +
ggalign(data = NULL, size = 0.2) +
geom_tile(aes(y = 1L, fill = .panel))
#> → heatmap built with `geom_tile()`
ggheatmap(matrix(rnorm(81), nrow = 9)) +
anno_top(size = 0.5) +
align_dendro(k = 3L) +
ggalign(data = NULL, size = 0.2) +
geom_tile(aes(y = 1L, fill = .panel))
#> → heatmap built with `geom_tile()`