The package offers a suite of align_* functions designed
to give you precise control over plot layout. These functions enable you
to manipulate axis order within the layout and partition along an axis
into multiple panels.
Currently, there are four key align_* functions
available for layout customization:
-
align_group: Group and align plots based on categorical factors. -
align_reorder: Reorder plots or split axes into different panels. -
align_kmeans: Arrange plots by k-means clustering results. -
align_dendro: Align plots according to hierarchical clustering or dendrograms.
set.seed(123)
small_mat <- matrix(rnorm(81), nrow = 9)
rownames(small_mat) <- paste0("row", seq_len(nrow(small_mat)))
colnames(small_mat) <- paste0("column", seq_len(ncol(small_mat)))
align_group
The align_group() function allows you to group
rows/columns into separate panels. It doesn’t add any plot area.
ggheatmap(small_mat) +
hmanno("t") +
align_group(sample(letters[1:4], ncol(small_mat), replace = TRUE))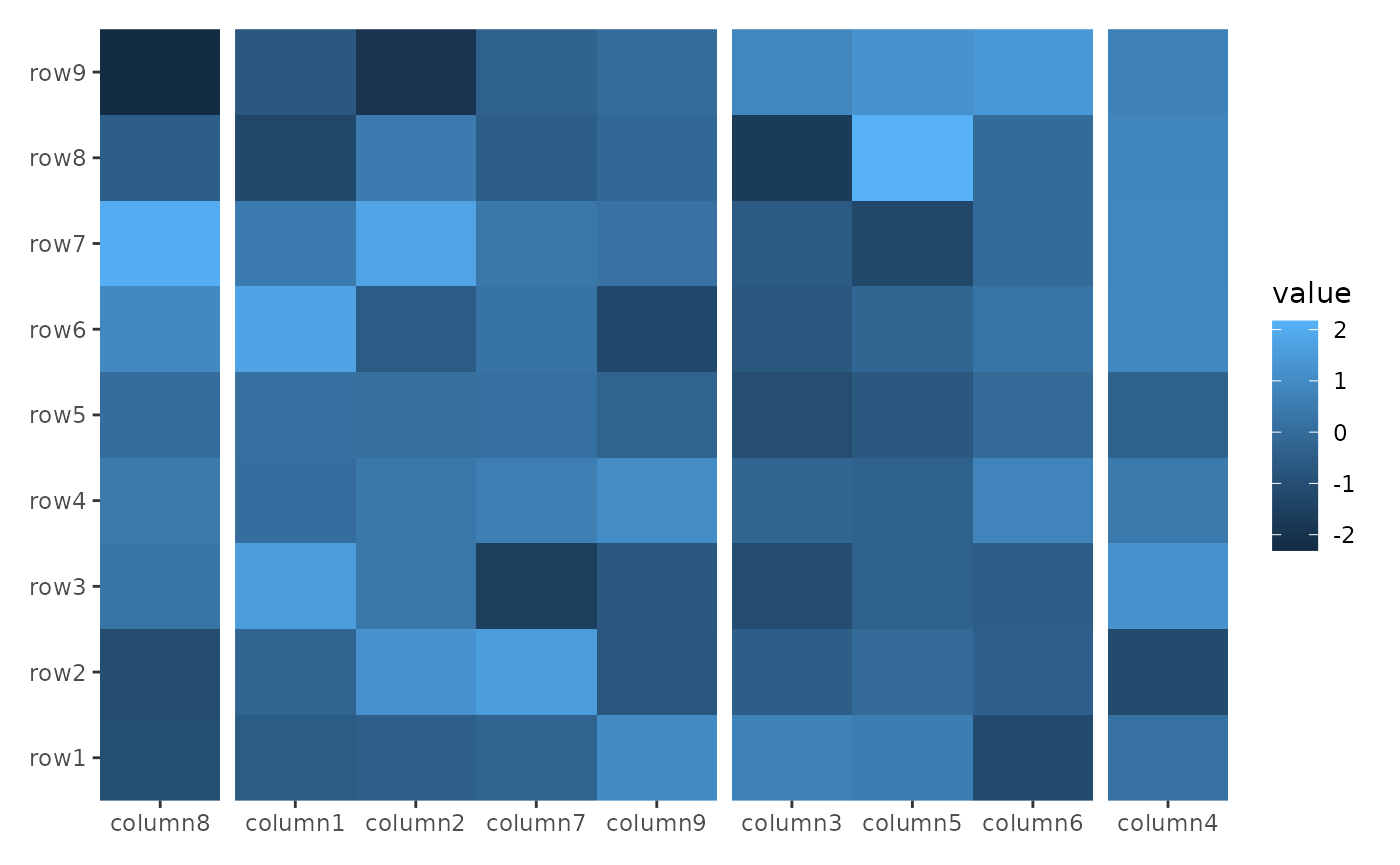
By default, the facet strip text is removed. You can override this
behavior with theme(strip.text = element_text()). Since
align_group() does not create a new plot, the panel title
can only be added to the heatmap plot.
ggheatmap(small_mat) +
theme(strip.text = element_text()) +
hmanno("l") +
align_group(sample(letters[1:4], nrow(small_mat), replace = TRUE))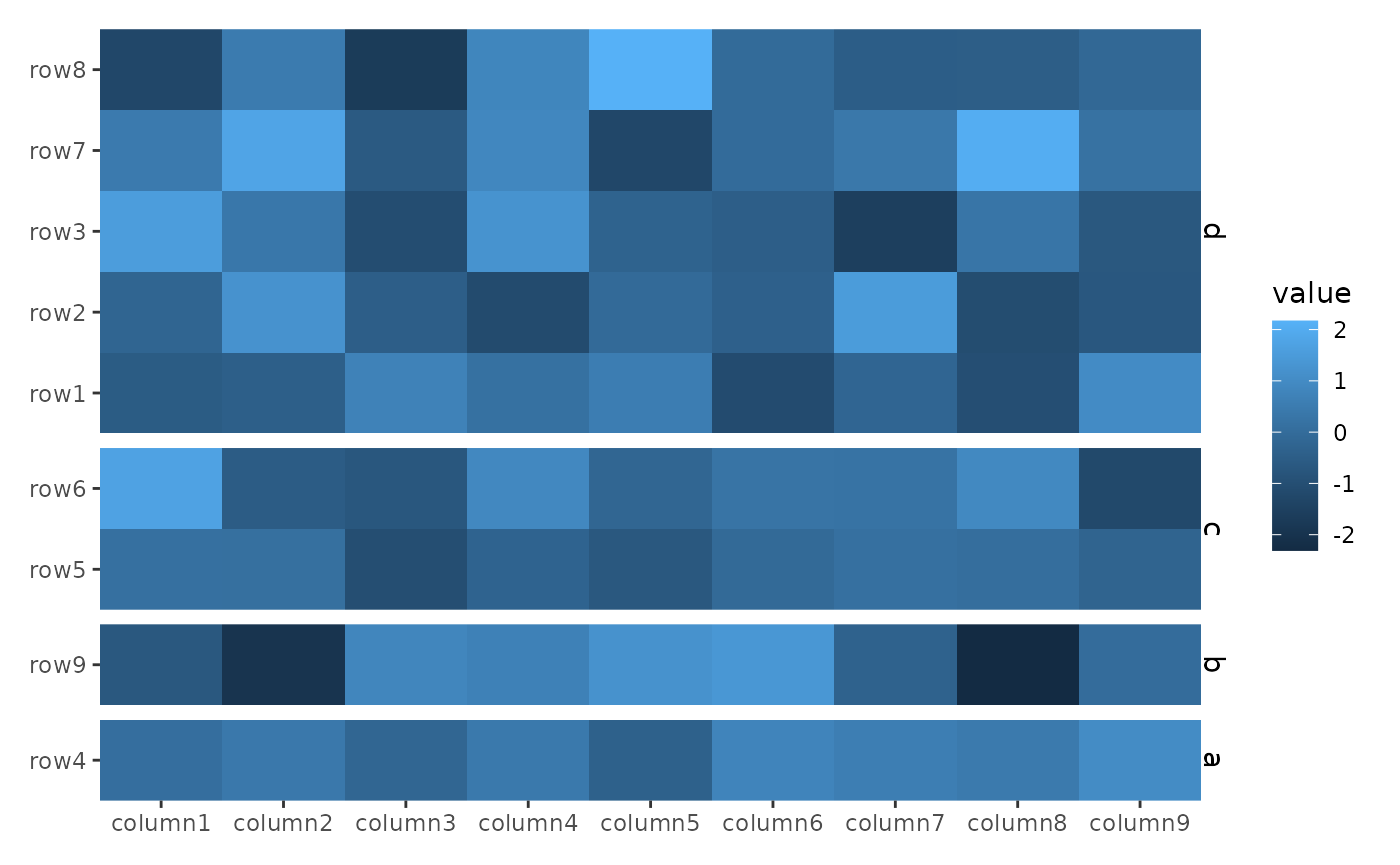
align_reorder
The align_reorder() function reorders the rows/columns
based on a summary function. Like align_group(), it doesn’t
add a plot area.
Here, we reorder the rows based on the means.
ggheatmap(small_mat) +
hmanno("l") +
align_reorder(rowMeans)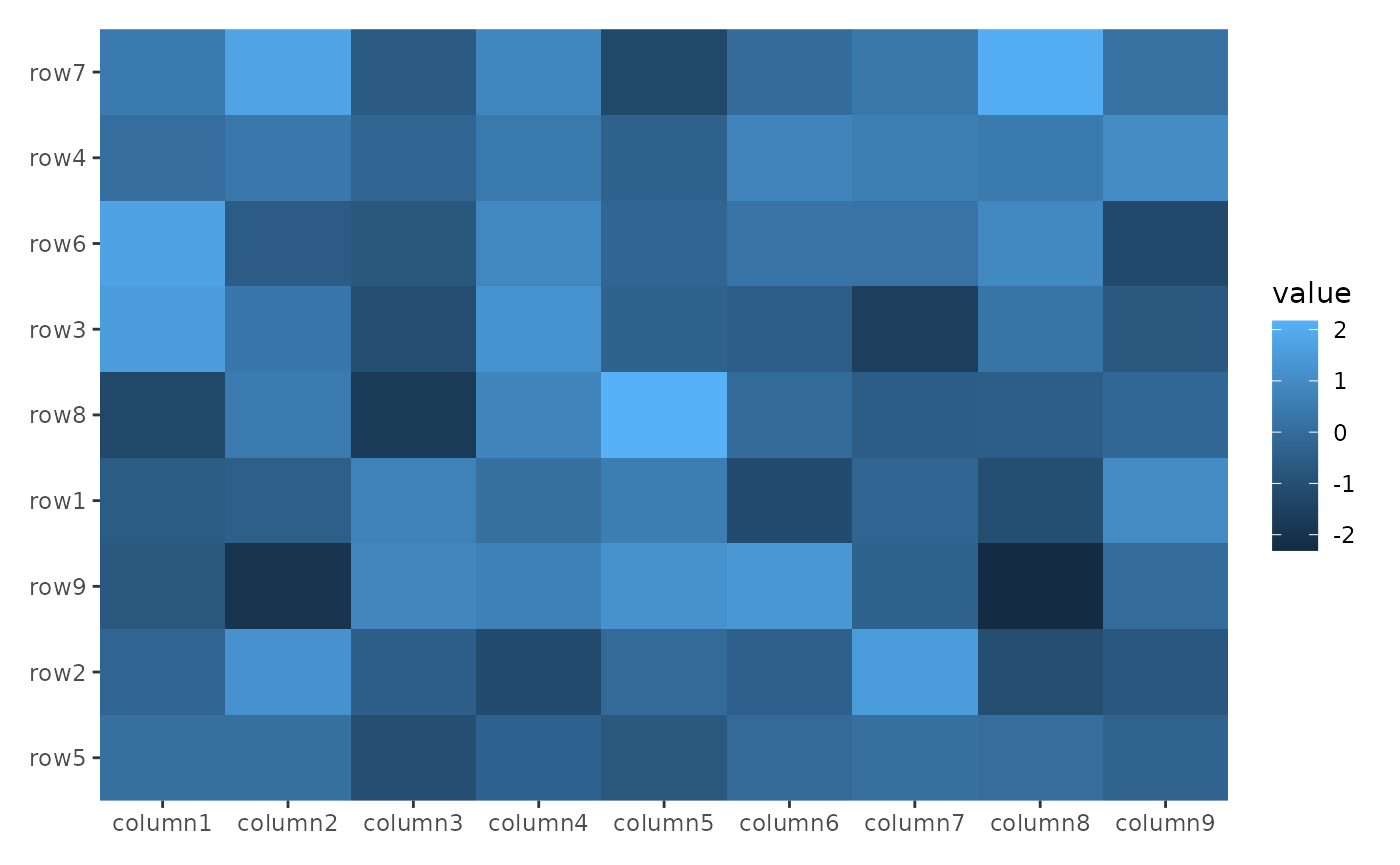
By default, align_reorder() reorders the rows or columns
in ascending order of the summary function’s output (from bottom to top
for rows, or from left to right for columns). To reverse this order, you
can set decreasing = TRUE:
ggheatmap(small_mat) +
hmanno("l") +
align_reorder(rowMeans, decreasing = TRUE)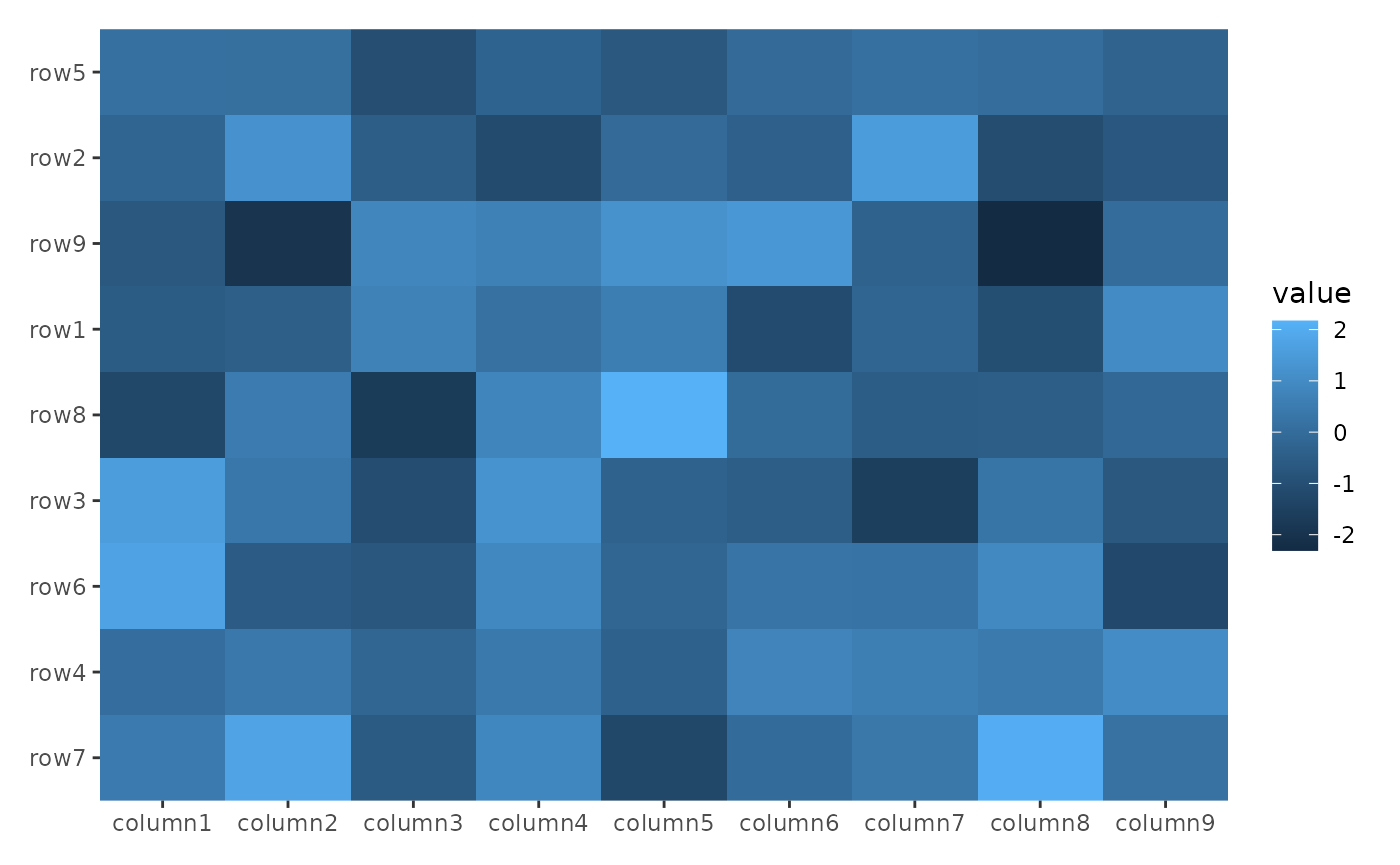
Some align_* functions accept a data
argument. This can be a matrix, a data frame, or even a simple vector,
which will be converted into a one-column matrix. If the
data argument is NULL, the function will use
the layout data, as demonstrated in the previous example. The
data argument can also accept a function (purrr-like lambda
syntax is supported), which will be applied to the layout data.
Note: All
align_*functions treat rows as observations, meaning thatNROW()function must return the same number as the parallel layout axis. For heatmap column annotations, the heatmap matrix is transposed before being used. Ifdatais a function, it will be applied to the transposed matrix.
Even for top and bottom annotations, you can use
rowMeans() to calculate the mean value across all
columns.
ggheatmap(small_mat) +
hmanno("t") +
align_reorder(rowMeans)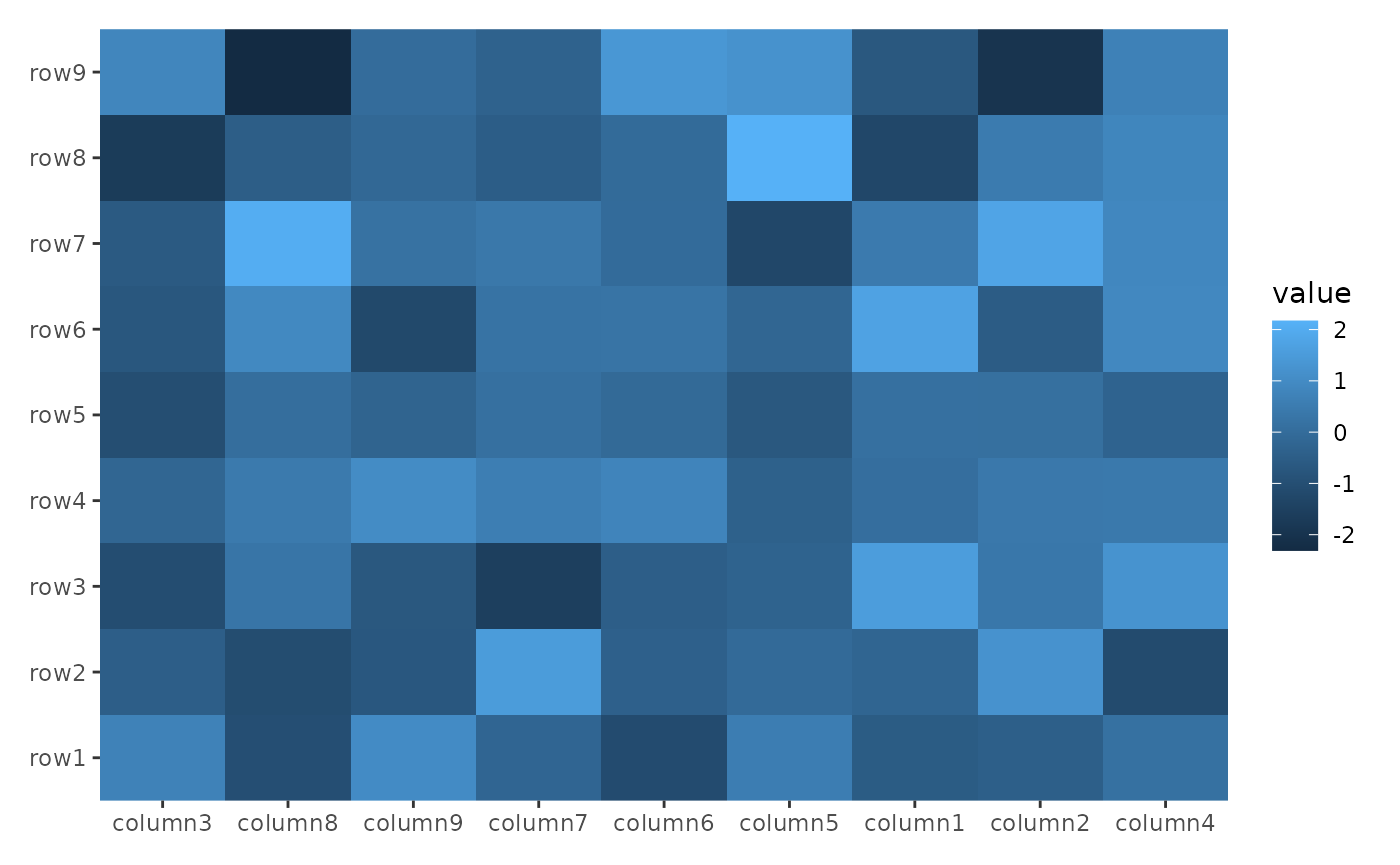
align_kmeans
The align_kmeans() function groups heatmap rows or
columns based on k-means clustering. Like the previous functions, it
does not add a plot area.
ggheatmap(small_mat) +
hmanno("t") +
align_kmeans(3L)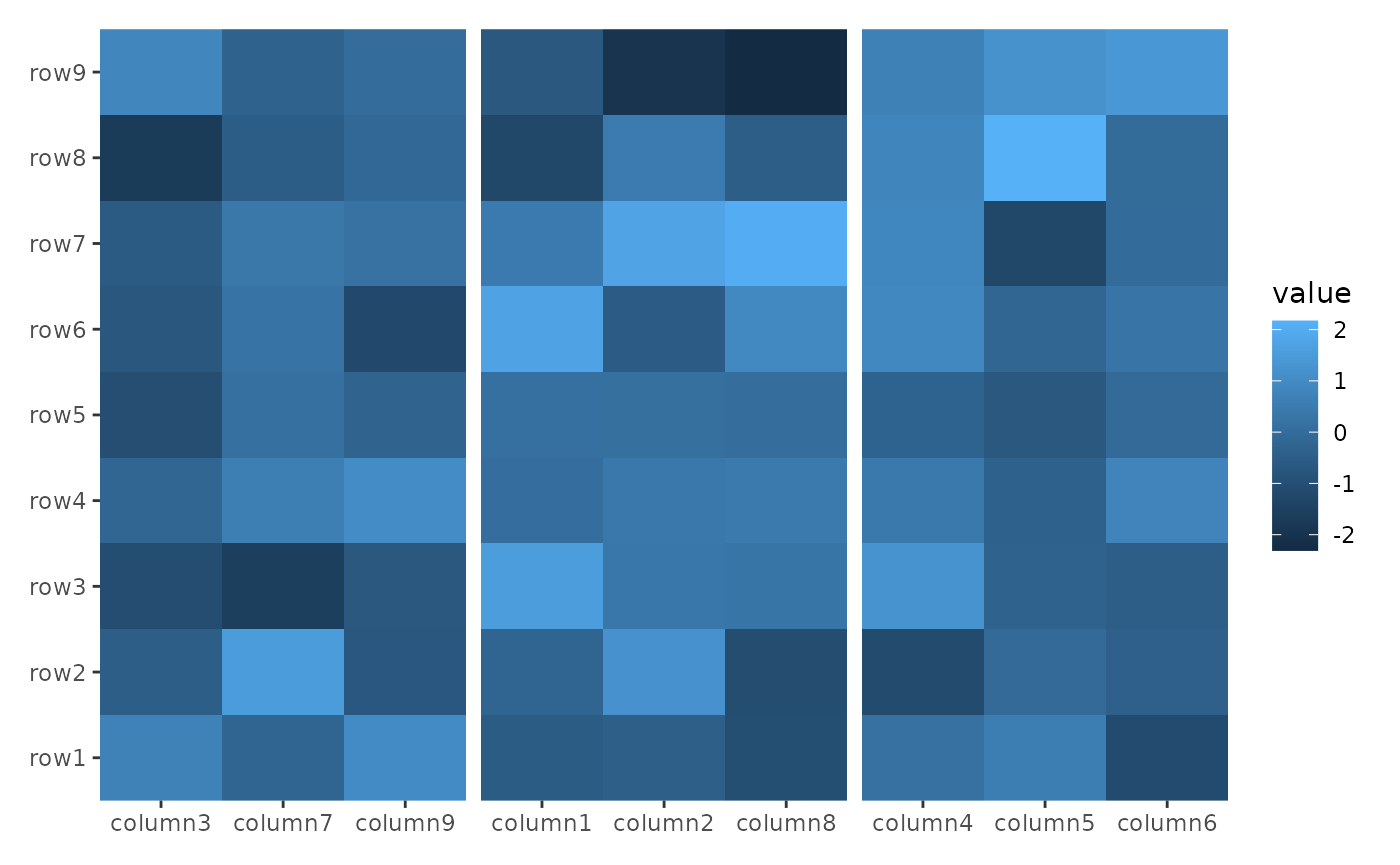
It’s important to note that align_group() and
align_kmeans() cannot do sub-grouping. This means they
cannot be used if groups already exist.
ggheatmap(small_mat) +
hmanno("t") +
align_group(sample(letters[1:4], ncol(small_mat), replace = TRUE)) +
align_kmeans(3L)
#> Error in `align_kmeans()`:
#> ! `align_kmeans()` cannot do sub-split
#> ℹ Group of layout x-axis already exists
ggheatmap(small_mat) +
hmanno("t") +
align_kmeans(3L) +
align_group(sample(letters[1:4], ncol(small_mat), replace = TRUE))
#> Error in `align_group()`:
#> ! `align_group()` cannot do sub-split
#> ℹ Group of layout x-axis already existsalign_dendro
The align_dendro() function adds a dendrogram to the
layout and can also reorder or split the layout based on hierarchical
clustering. This is particularly useful for working with heatmap
plots.
ggheatmap(small_mat) +
hmanno("t") +
align_dendro()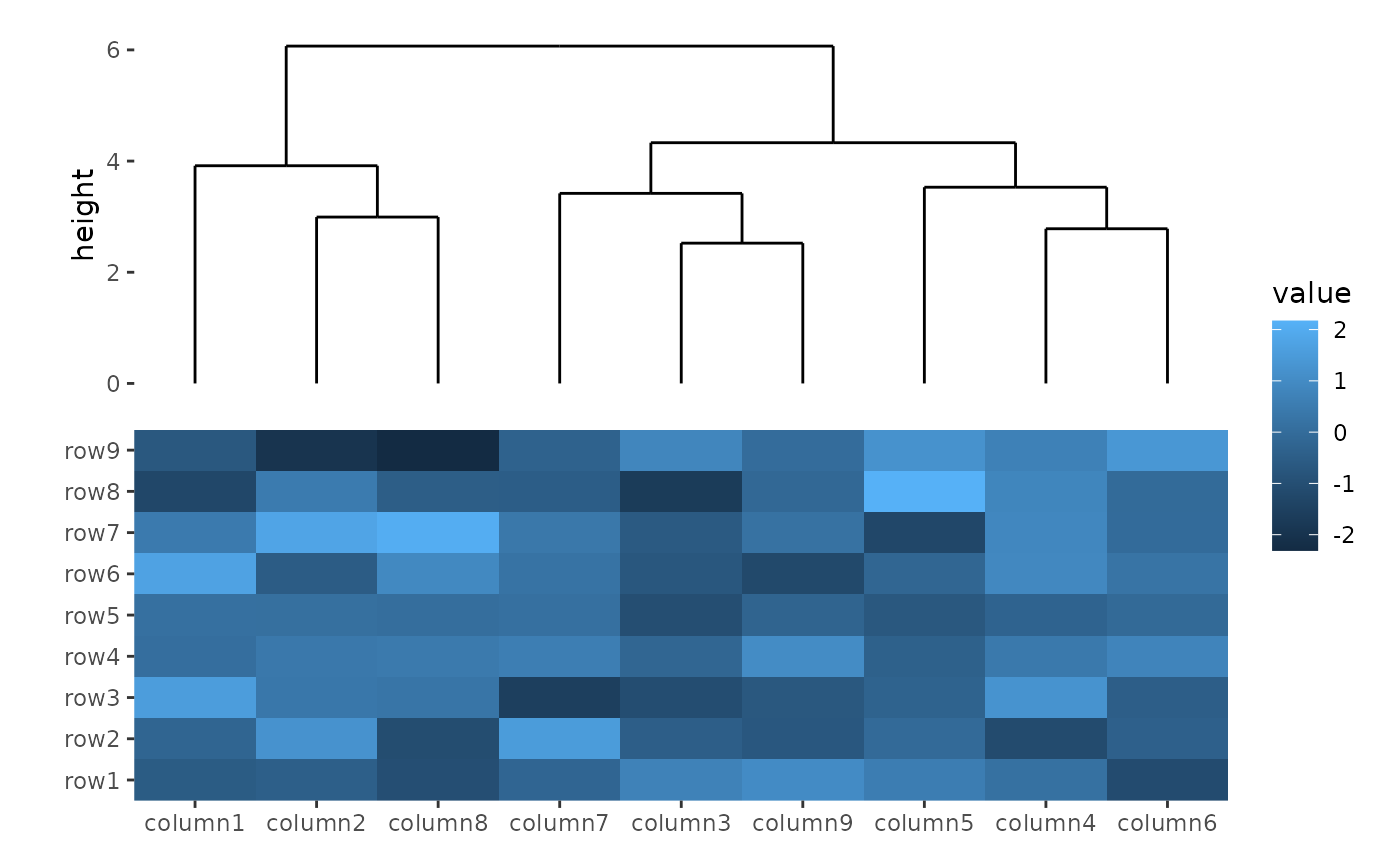
Hierarchical clustering is performed in two steps: calculate the
distance matrix and apply clustering. You can use the
distance and method argument to control the
dendrogram builind process.
There are two ways to specify distance metric for
clustering:
- specify
distanceas a pre-defined option. The valid values are the supported methods indist()function and coorelation coefficient"pearson","spearman"and"kendall". The correlation distance is defined as1 - cor(x, y, method = distance). - a self-defined function which calculates distance from a matrix. The function should only contain one argument. Please note for clustering on columns, the matrix will be transposed automatically.
ggheatmap(small_mat) +
hmanno("t") +
align_dendro(distance = "pearson") +
patch_titles(top = "pre-defined distance method (1 - pearson)")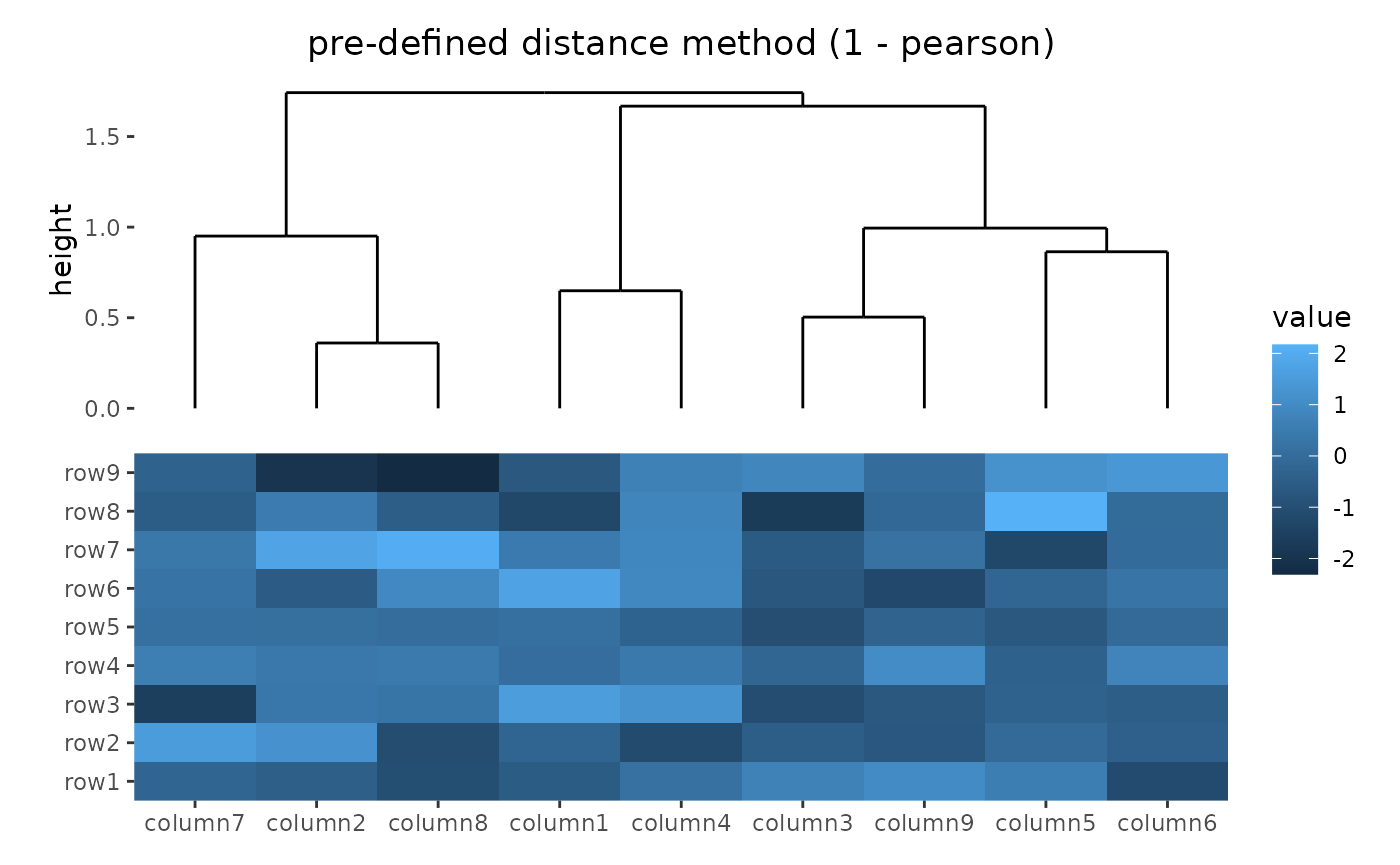
ggheatmap(small_mat) +
hmanno("t") +
align_dendro(distance = function(m) dist(m)) +
patch_titles(top = "a function that calculates distance matrix")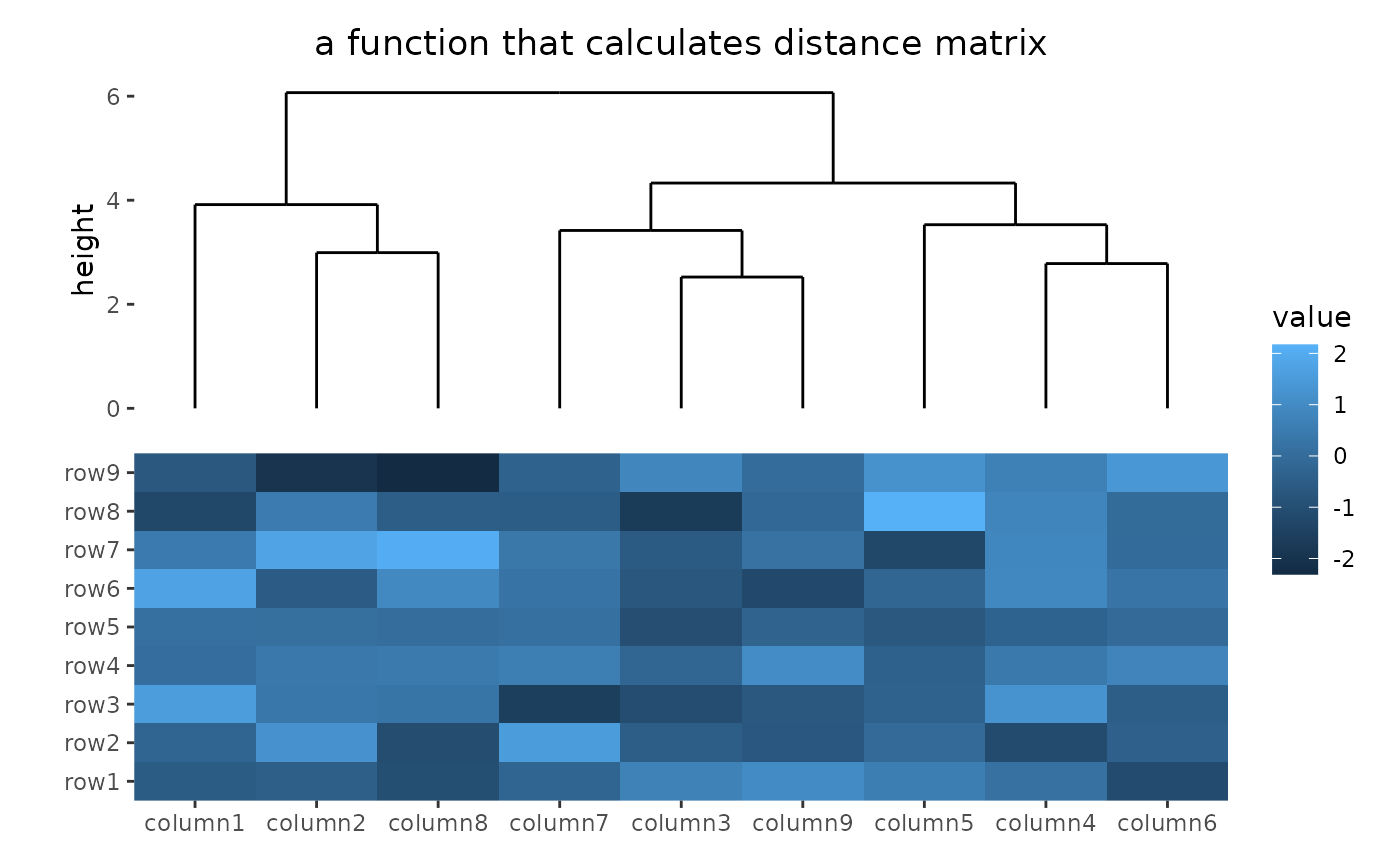
Method to perform hierarchical clustering can be specified by
method. Possible methods are those supported in
hclust() function. And you can also provide a self-defined
function, which accepts the distance object and return a
hclust object.
ggheatmap(small_mat) +
hmanno("t") +
align_dendro(method = "ward.D2")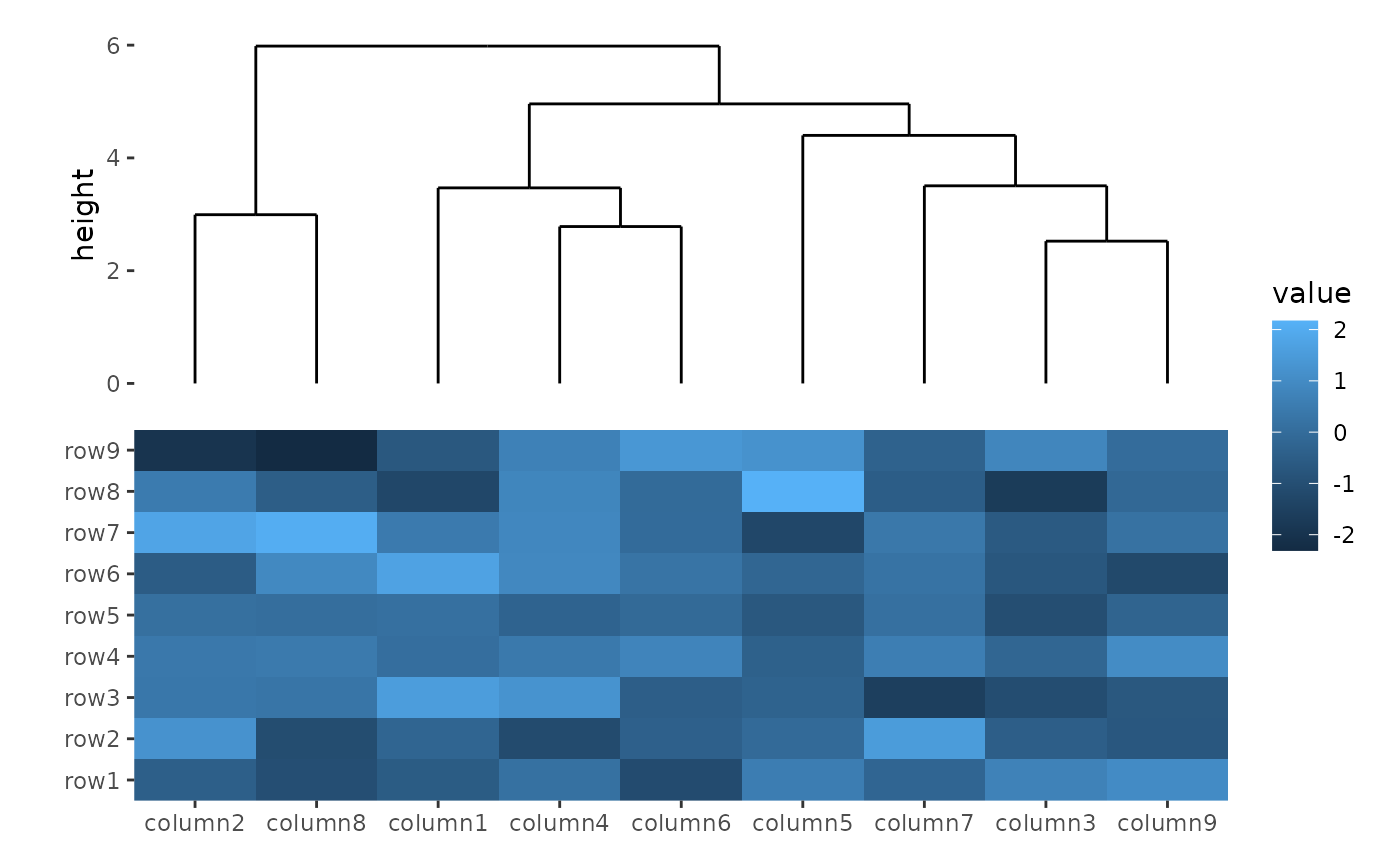
The dendrogram can also be used to cut the columns/rows into groups.
You can specify k or h, which work similarly
to cutree():
ggheatmap(small_mat) +
hmanno("t") +
align_dendro(k = 3L)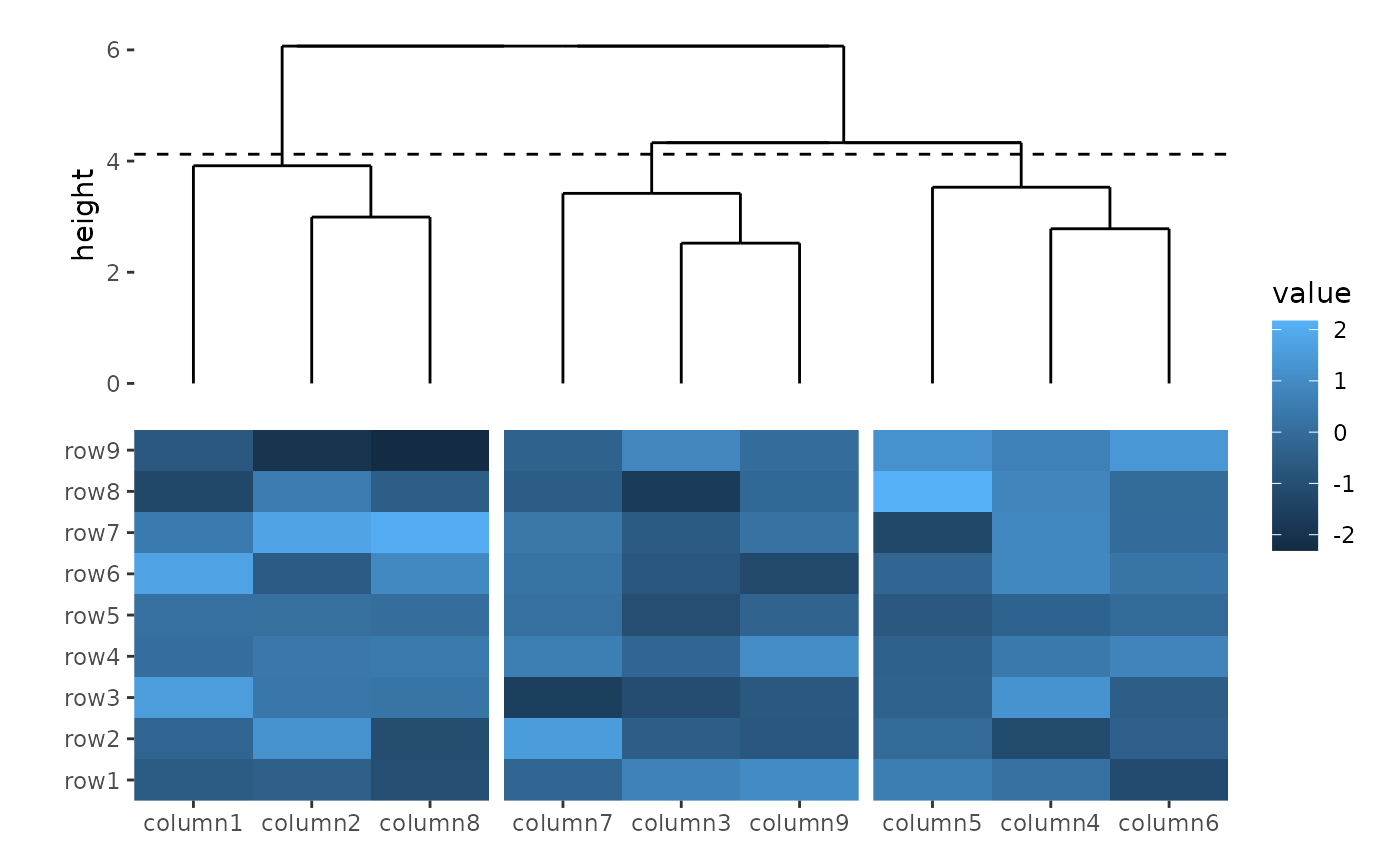
In contrast to align_group(),
align_kmeans(), and align_reorder(),
align_dendro() is capable of drawing plot components. So it
has a default set_context value of TRUE,
meaning it will set the active context of the annotation stack layout.
In this way, we can add any ggplot elements to this plot area.
ggheatmap(small_mat) +
hmanno("t") +
align_dendro() +
geom_point(aes(y = y))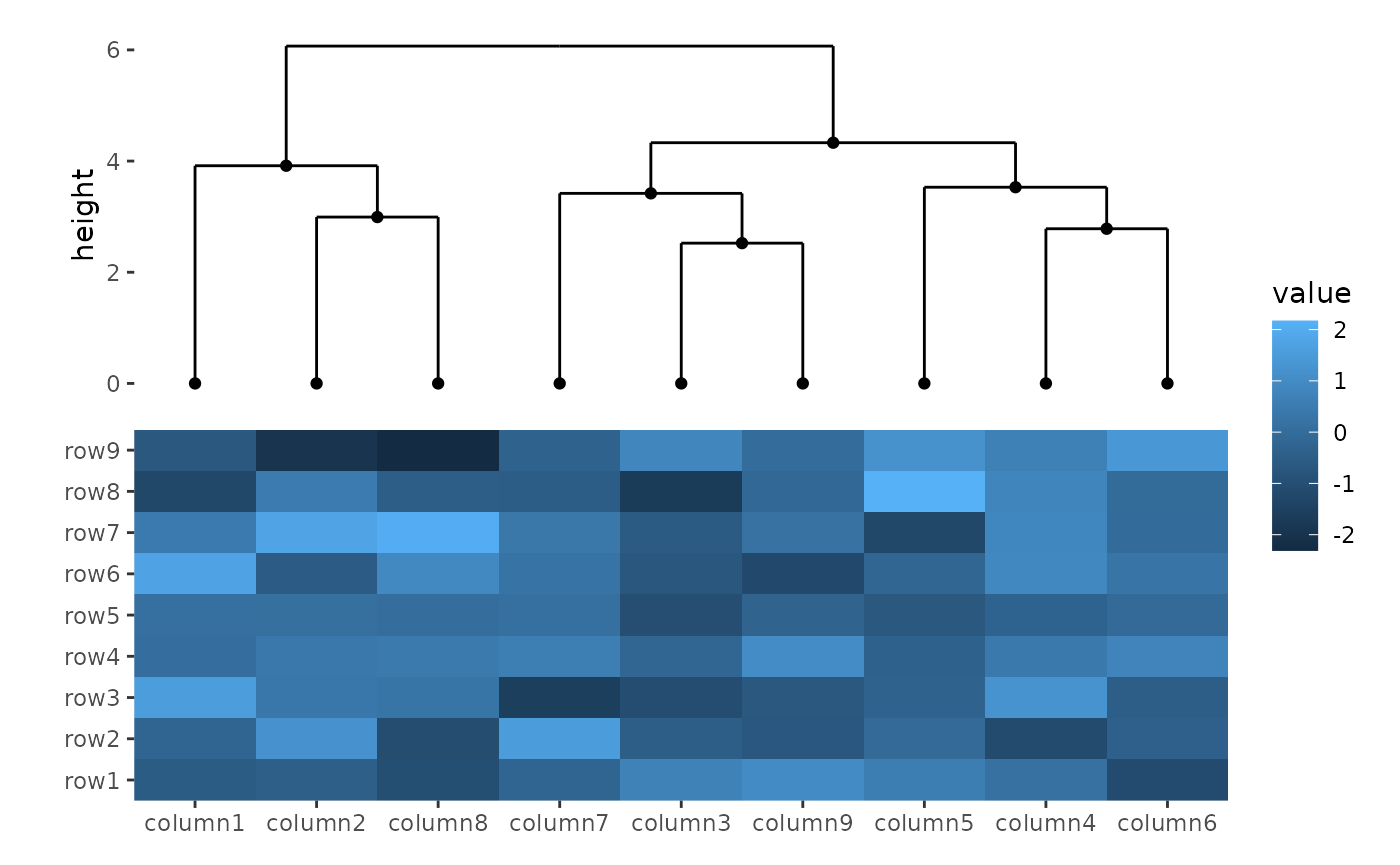
The align_dendro() function creates default
node data for the ggplot. See
ggplot2 specification in ?align_dendro for
details. Additionally, edge data is added to the
ggplote::geom_segment() layer directly, used to draw the
dendrogram tree. One useful variable in both node and
edge data is the branch column, corresponding
to the cutree result:
ggheatmap(small_mat) +
hmanno("t") +
align_dendro(aes(color = branch), k = 3) +
geom_point(aes(color = branch, y = y))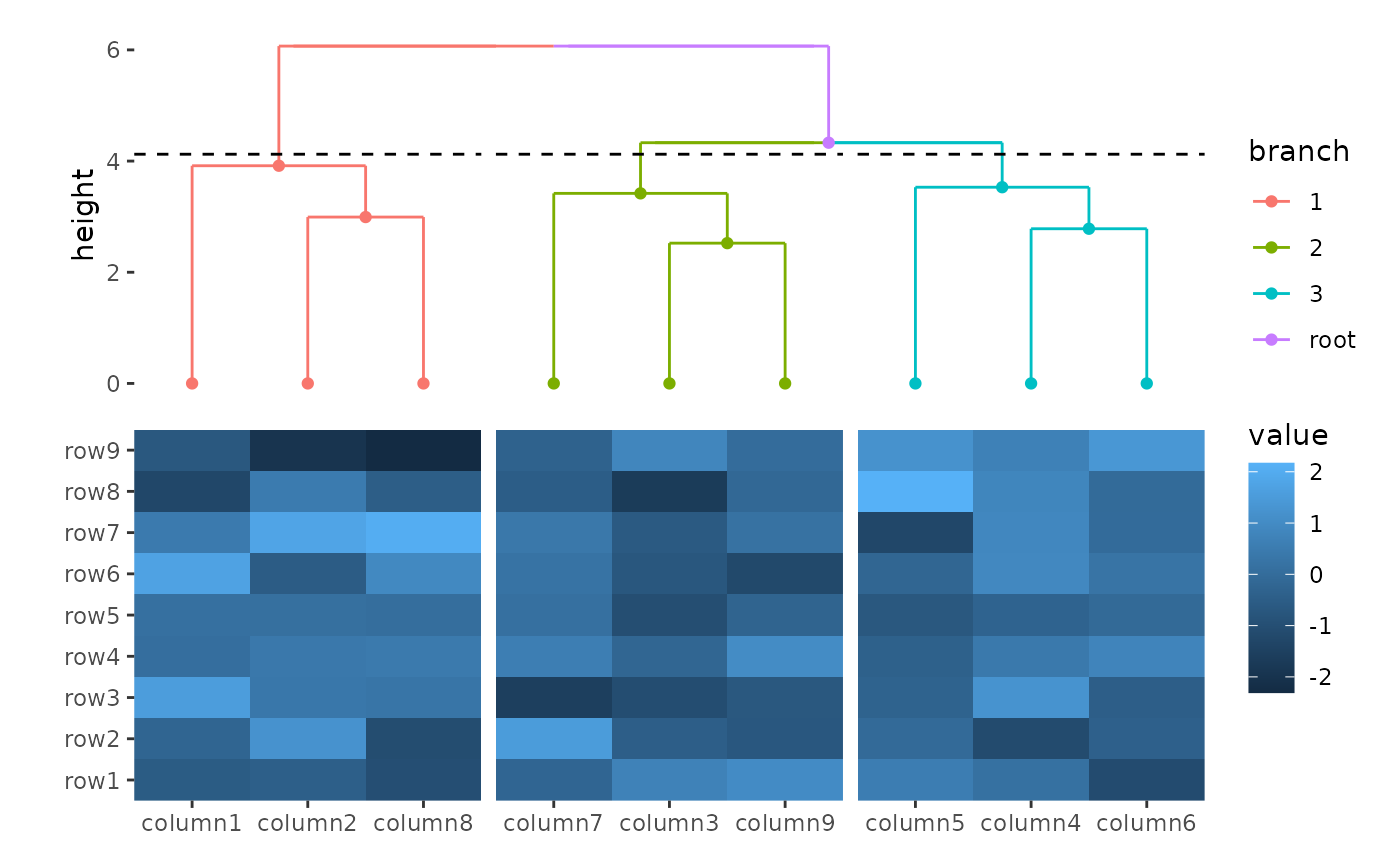
align_dendro() can also perform clustering between
groups, meaning it can be used even if there are existing groups present
in the layout:
column_groups <- sample(letters[1:3], ncol(small_mat), replace = TRUE)
ggheatmap(small_mat) +
hmanno("t") +
align_group(column_groups) +
align_dendro(aes(color = branch))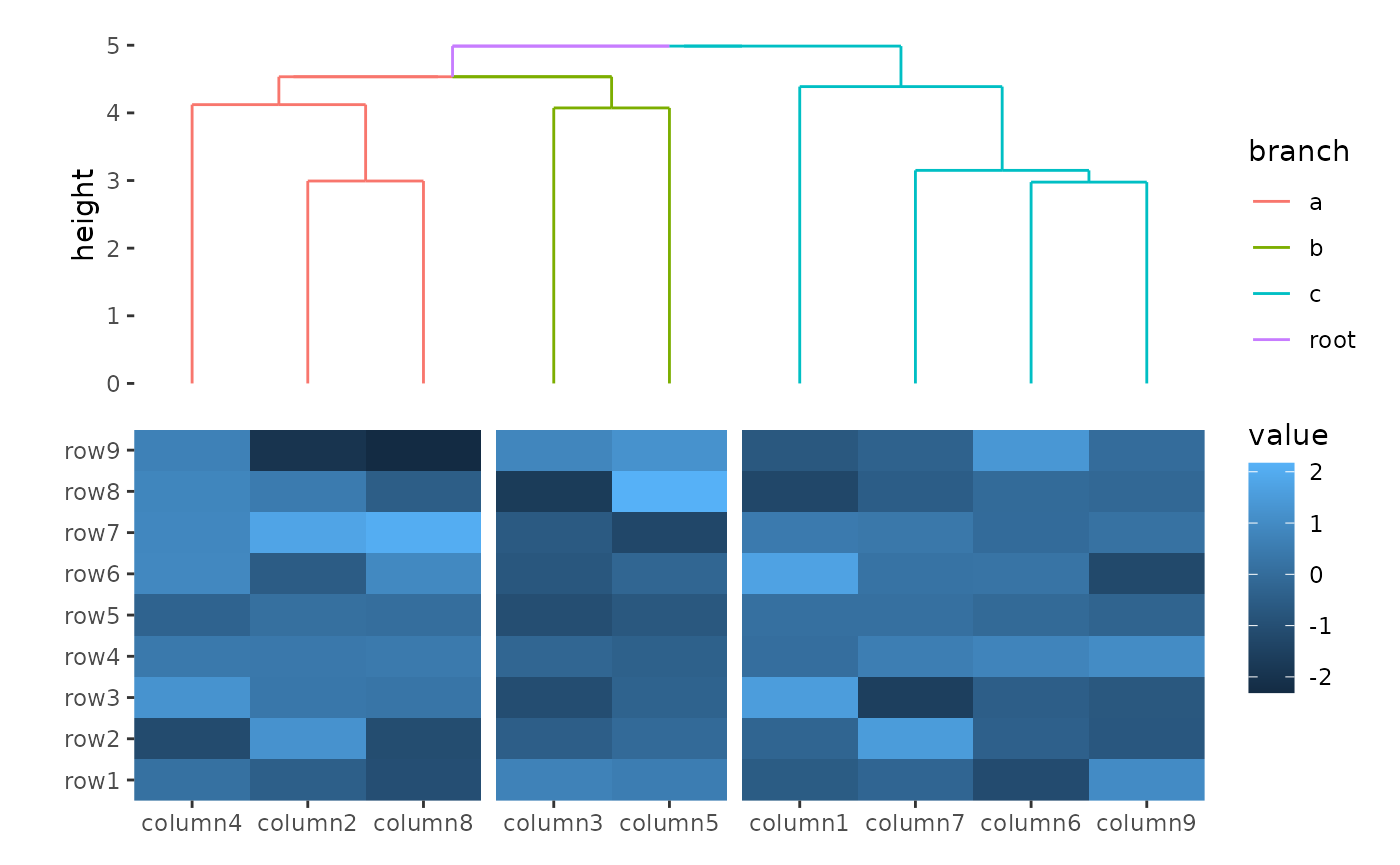
You can reorder the groups by setting
reorder_group = TRUE.
ggheatmap(small_mat) +
hmanno("t") +
align_group(column_groups) +
align_dendro(aes(color = branch), reorder_group = TRUE)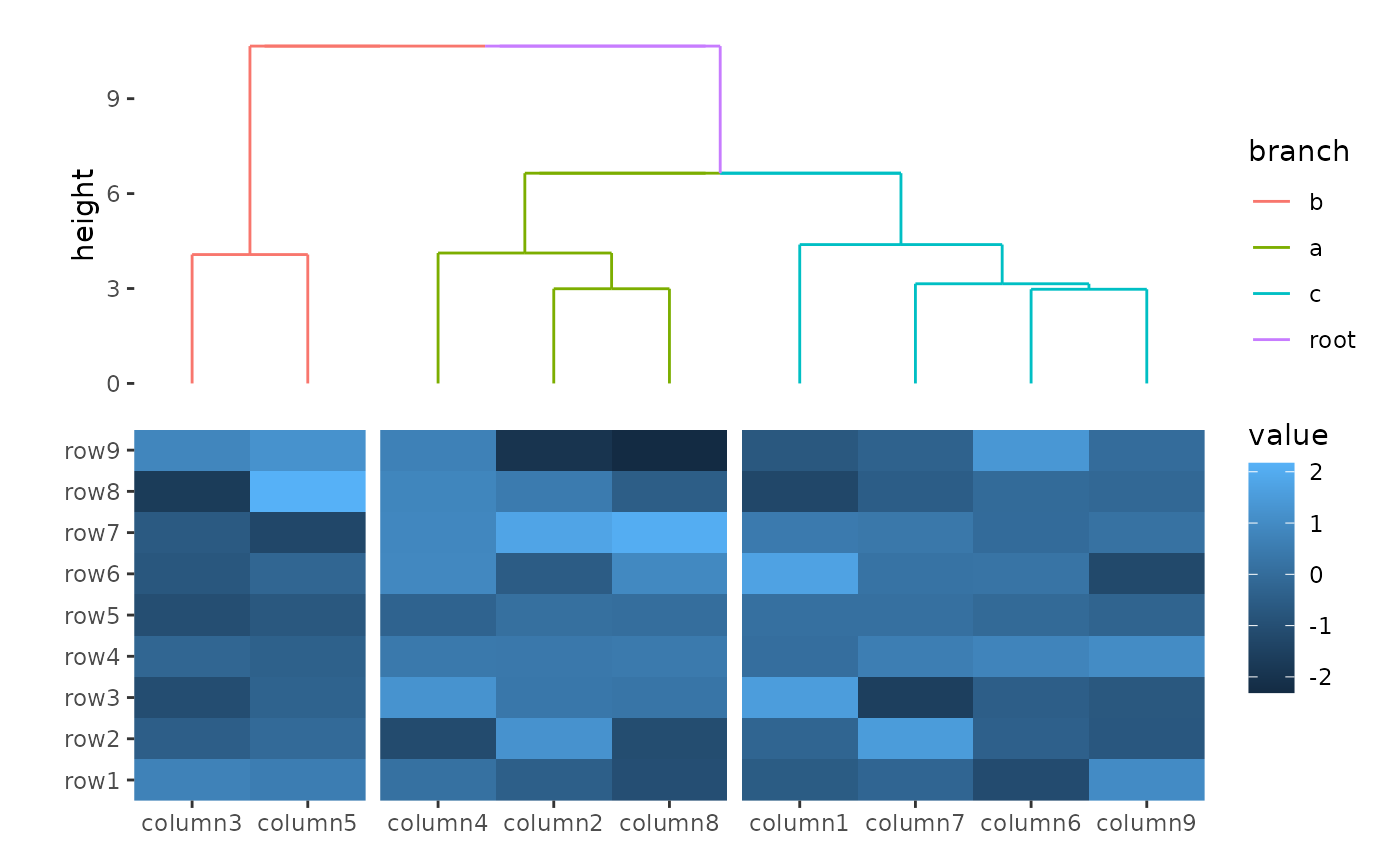
You can see the difference by drawing two dendrogram.
ggheatmap(small_mat) +
hmanno("t") +
align_group(column_groups) +
align_dendro(aes(color = branch), reorder_group = TRUE) +
hmanno("b") +
align_dendro(aes(color = branch), reorder_group = FALSE)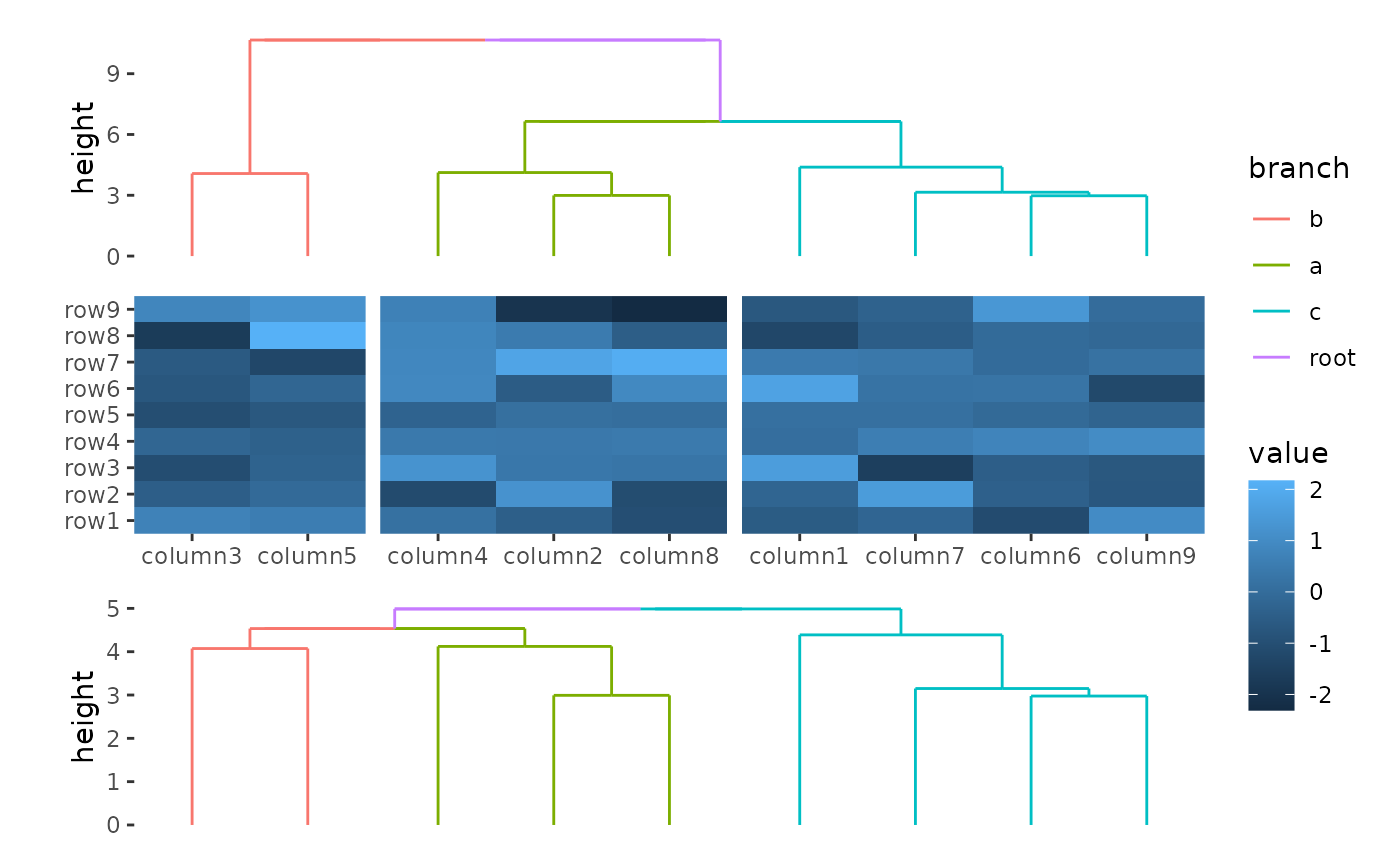
Session information
sessionInfo()
#> R version 4.4.1 (2024-06-14)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggalign_0.0.4 ggplot2_3.5.1
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.5 jsonlite_1.8.8 highr_0.11 dplyr_1.1.4
#> [5] compiler_4.4.1 tidyselect_1.2.1 tidyr_1.3.1 jquerylib_0.1.4
#> [9] systemfonts_1.1.0 scales_1.3.0 textshaping_0.4.0 yaml_2.3.10
#> [13] fastmap_1.2.0 ggh4x_0.2.8 R6_2.5.1 labeling_0.4.3
#> [17] generics_0.1.3 knitr_1.48 tibble_3.2.1 desc_1.4.3
#> [21] munsell_0.5.1 bslib_0.8.0 pillar_1.9.0 rlang_1.1.4
#> [25] utf8_1.2.4 cachem_1.1.0 xfun_0.47 fs_1.6.4
#> [29] sass_0.4.9 cli_3.6.3 pkgdown_2.1.1 withr_3.0.1
#> [33] magrittr_2.0.3 digest_0.6.37 grid_4.4.1 lifecycle_1.0.4
#> [37] vctrs_0.6.5 evaluate_1.0.0 glue_1.7.0 farver_2.1.2
#> [41] ragg_1.3.3 fansi_1.0.6 colorspace_2.1-1 rmarkdown_2.28
#> [45] purrr_1.0.2 tools_4.4.1 pkgconfig_2.0.3 htmltools_0.5.8.1