In this thread, we’ll use ggalign to draw all the
heatmap in https://jokergoo.github.io/ComplexHeatmap-reference/book/a-single-heatmap.html
set.seed(123)
nr1 <- 4
nr2 <- 8
nr3 <- 6
nr <- nr1 + nr2 + nr3
nc1 <- 6
nc2 <- 8
nc3 <- 10
nc <- nc1 + nc2 + nc3
mat <- cbind(
rbind(
matrix(rnorm(nr1 * nc1, mean = 1, sd = 0.5), nrow = nr1),
matrix(rnorm(nr2 * nc1, mean = 0, sd = 0.5), nrow = nr2),
matrix(rnorm(nr3 * nc1, mean = 0, sd = 0.5), nrow = nr3)
),
rbind(
matrix(rnorm(nr1 * nc2, mean = 0, sd = 0.5), nrow = nr1),
matrix(rnorm(nr2 * nc2, mean = 1, sd = 0.5), nrow = nr2),
matrix(rnorm(nr3 * nc2, mean = 0, sd = 0.5), nrow = nr3)
),
rbind(
matrix(rnorm(nr1 * nc3, mean = 0.5, sd = 0.5), nrow = nr1),
matrix(rnorm(nr2 * nc3, mean = 0.5, sd = 0.5), nrow = nr2),
matrix(rnorm(nr3 * nc3, mean = 1, sd = 0.5), nrow = nr3)
)
)
mat <- mat[sample(nr, nr), sample(nc, nc)]
rownames(mat) <- paste0("row", seq_len(nr))
colnames(mat) <- paste0("column", seq_len(nc))Colors
It is important to note that the ComplexHeatmap package
reorders the dendrogram by default, while align_dendro() in
ggalign does not modify the tree layout.
Another key difference is in how the two packages treat the starting
point. ggalign considers the left-bottom as the starting
point, whereas ComplexHeatmap starts from the left-top.
When reordering the dendrogram, ComplexHeatmap does so in
decreasing order, while ggalign uses an ascending
order.
To modify colors in the heatmap, you can use the
scale_fill_*() function from ggplot2, which
provides a flexible way and enriched pallete to adjust color
schemes.
# ComplexHeatmap::Heatmap(mat)
dim(mat)
#> [1] 18 24
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`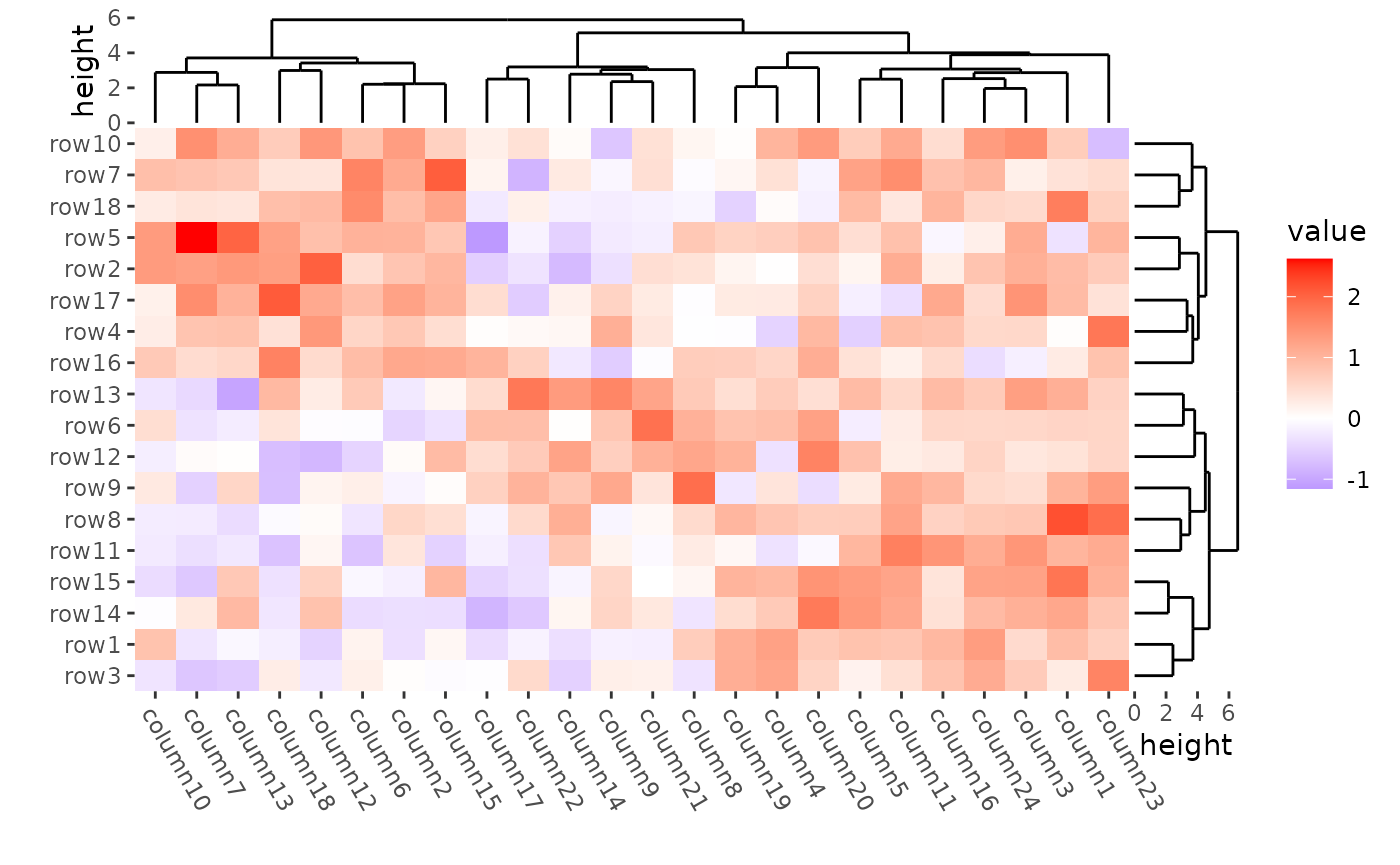
ggheatmap(mat) +
scale_fill_gradient2(low = "green", high = "red") +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro() +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`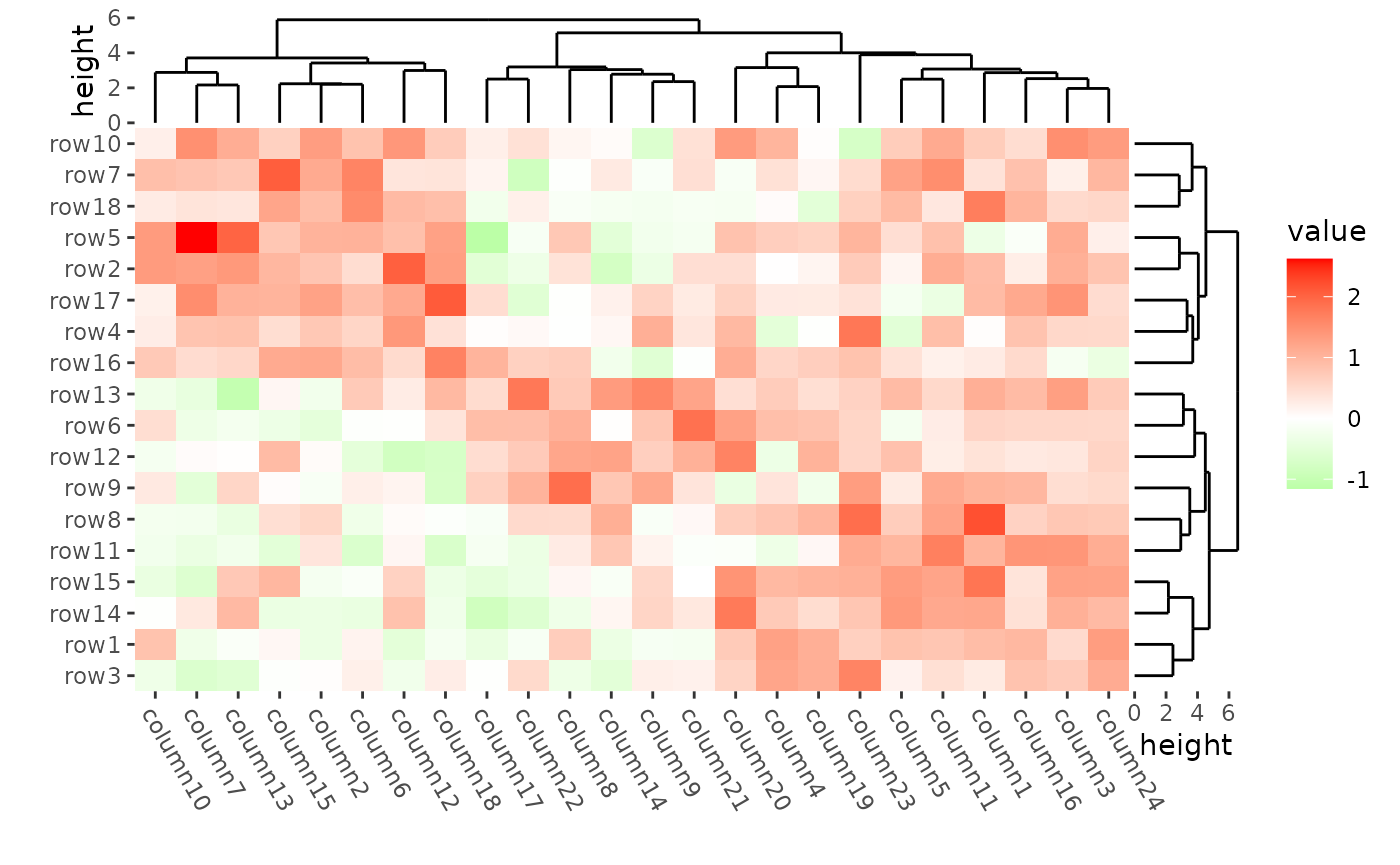
oob argument in the scale_fill_* function
can be used to deal with the outliers.
mat2 <- mat
mat2[1, 1] <- 100000
ggheatmap(mat2) +
scale_fill_gradient2(
low = "green", high = "red",
limits = c(-2, 2),
oob = scales::squish
) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`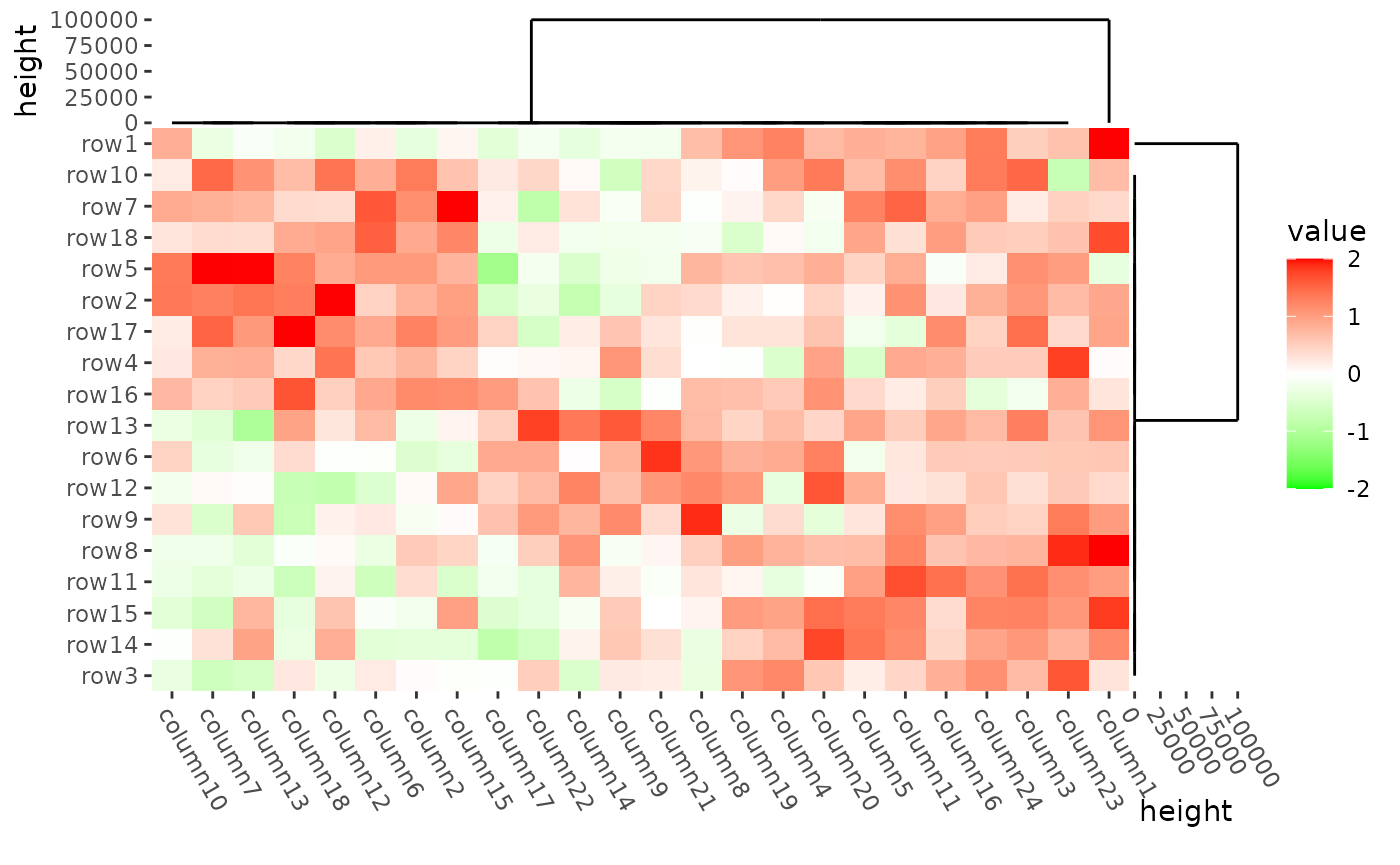
We can use align_plots() to arrange them.
h1 <- ggheatmap(mat) +
scale_fill_gradient2(name = "mat", low = "green", high = "red") +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
h2 <- ggheatmap(mat / 4) +
scale_fill_gradient2(
name = "mat/4", limits = c(-2, 2L),
oob = scales::squish,
low = "green", high = "red"
) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
h3 <- ggheatmap(abs(mat)) +
scale_fill_gradient2(name = "abs(mat)", low = "green", high = "red") +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
align_plots(h1, h2, h3, ncol = 2L)
#> → heatmap built with `geom_tile()`
#> → heatmap built with `geom_tile()`
#> → heatmap built with `geom_tile()`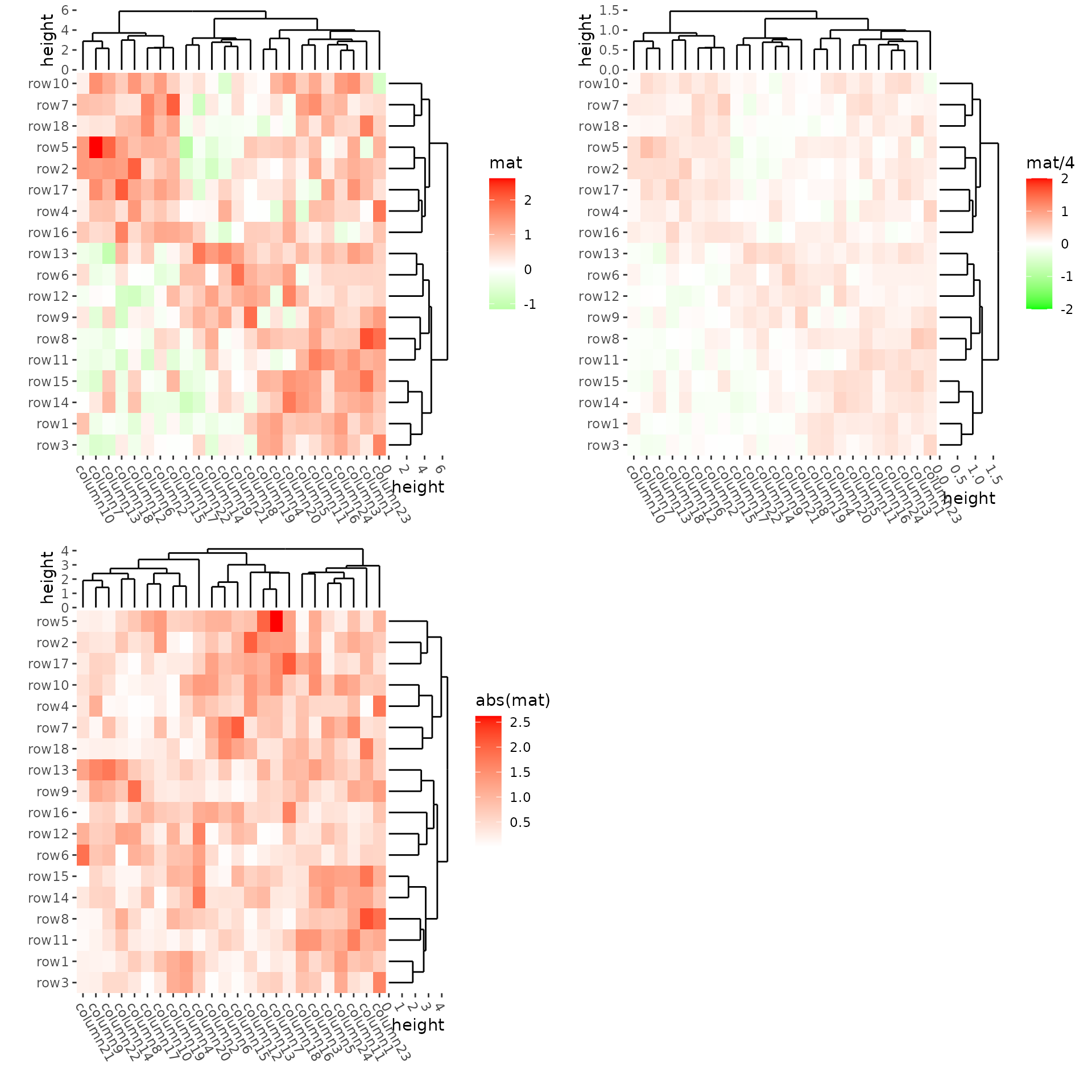
ggheatmap(mat) +
scale_fill_gradientn(colors = rev(rainbow(10))) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`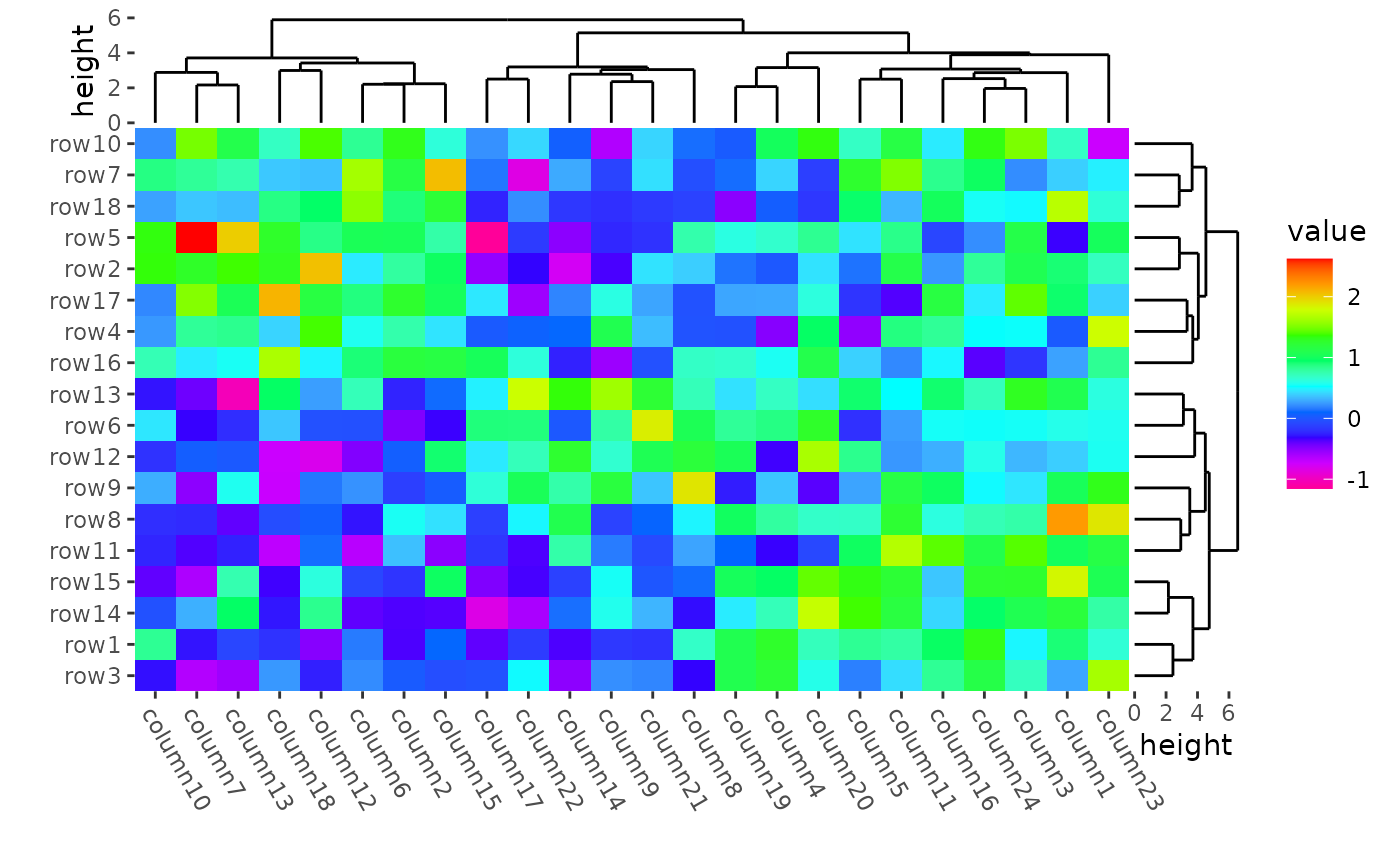
For character matrix, you can use ggplot2 discrete filling scales.
discrete_mat <- matrix(sample(1:4, 100, replace = TRUE), 10, 10)
colors <- structure(1:4, names = c("1", "2", "3", "4")) # black, red, green, blue
ggheatmap(discrete_mat, aes(fill = factor(value))) +
scale_fill_manual(values = colors) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`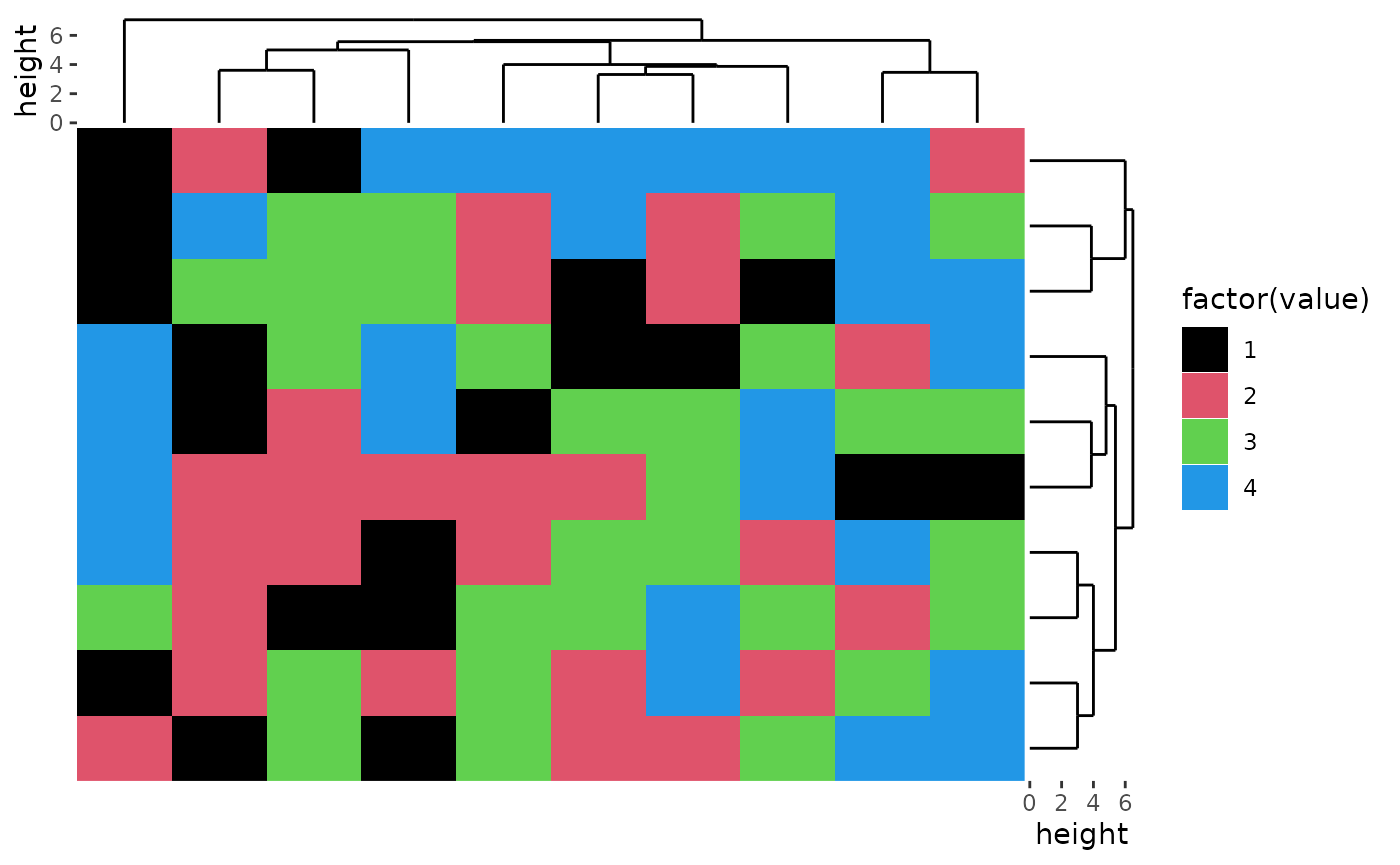
discrete_mat <- matrix(sample(letters[1:4], 100, replace = TRUE), 10, 10)
colors <- structure(1:4, names = letters[1:4])
ggheatmap(discrete_mat) +
scale_fill_manual(values = colors)
#> → heatmap built with `geom_tile()`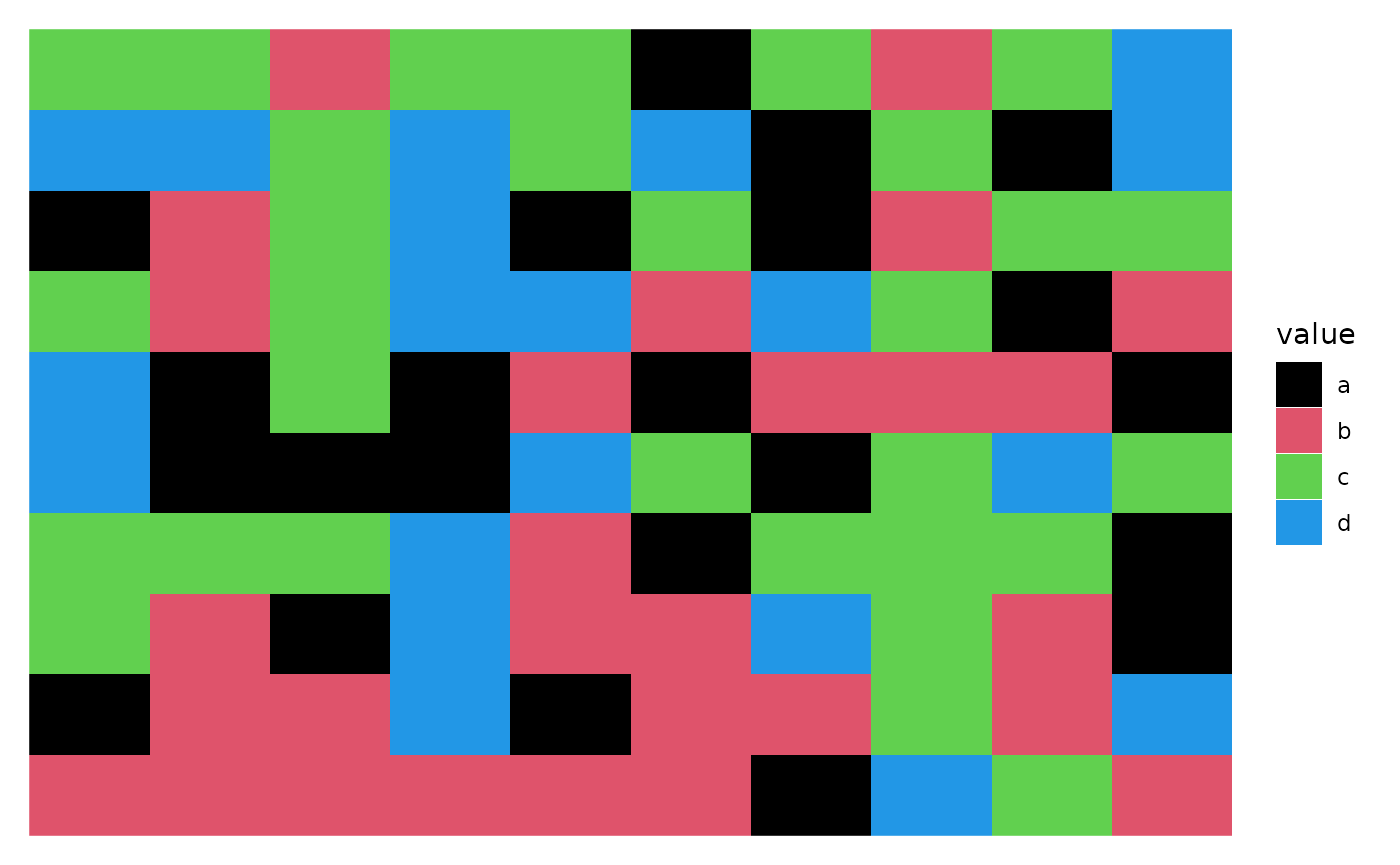
mat_with_na <- mat
na_index <- sample(c(TRUE, FALSE),
nrow(mat) * ncol(mat),
replace = TRUE, prob = c(1, 9)
)
mat_with_na[na_index] <- NA
ggheatmap(mat_with_na) +
scale_fill_gradient2(low = "blue", high = "red", na.value = "black") +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`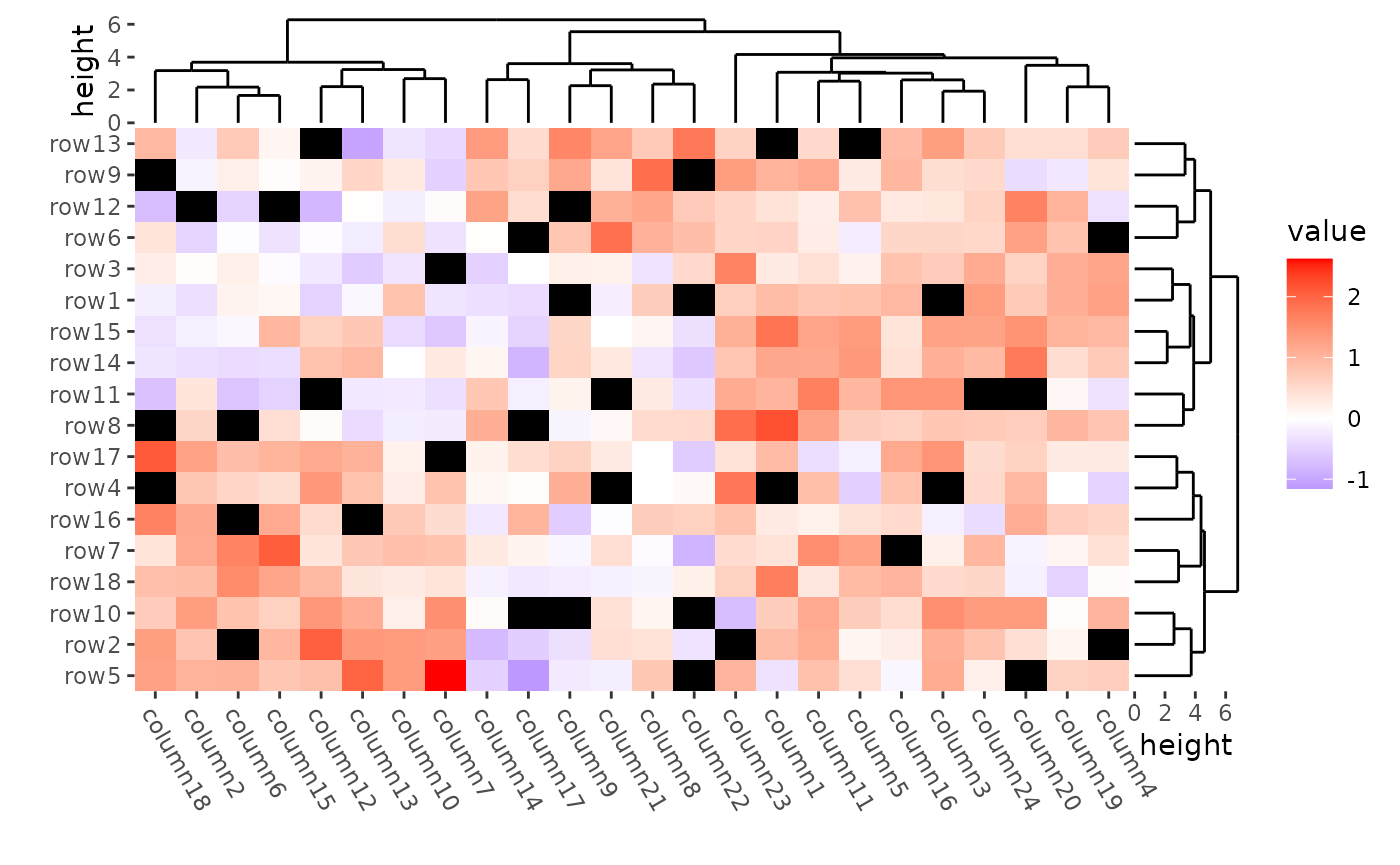
We won’t compare the LAB and RGB space. If you want to convert color between different color space, try to use farver pacakge.
In ggplot2, you can use panel.border argument in
theme() function to control the Heatmap body border.
ggheatmap(mat) +
theme(
axis.text.x = element_text(angle = -60, hjust = 0),
panel.border = element_rect(linetype = "dashed", fill = NA)
) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`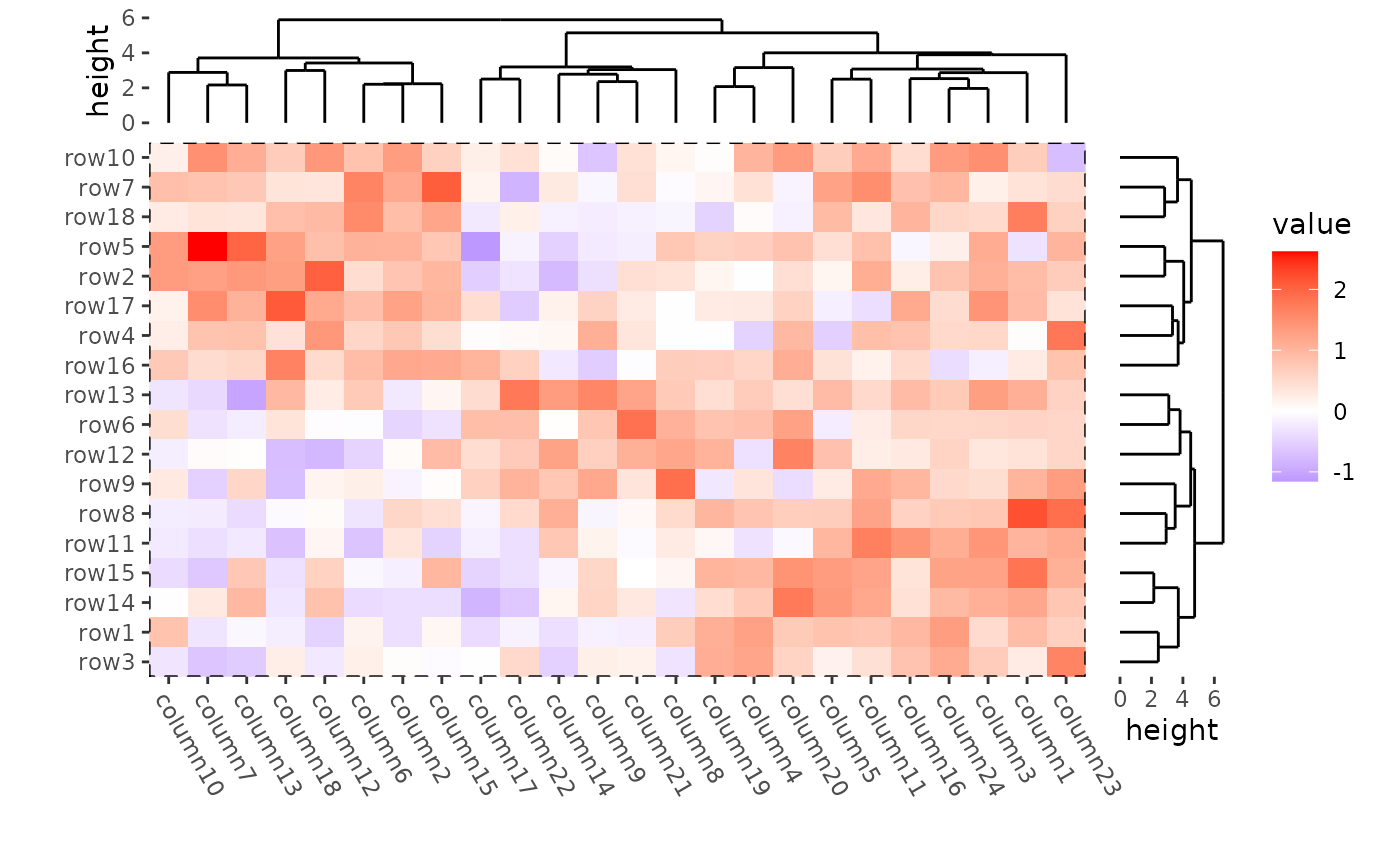
You can use the filling argument to turn off the heatmap
cell filling, allowing you to customize the heatmap body geoms. Use the
color aesthetic to specify the cell border color and the
linewidth aesthetic to set the border width.
ggheatmap(mat, filling = NULL) +
geom_tile(aes(fill = value), color = "white") +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) &
theme(plot.margin = margin())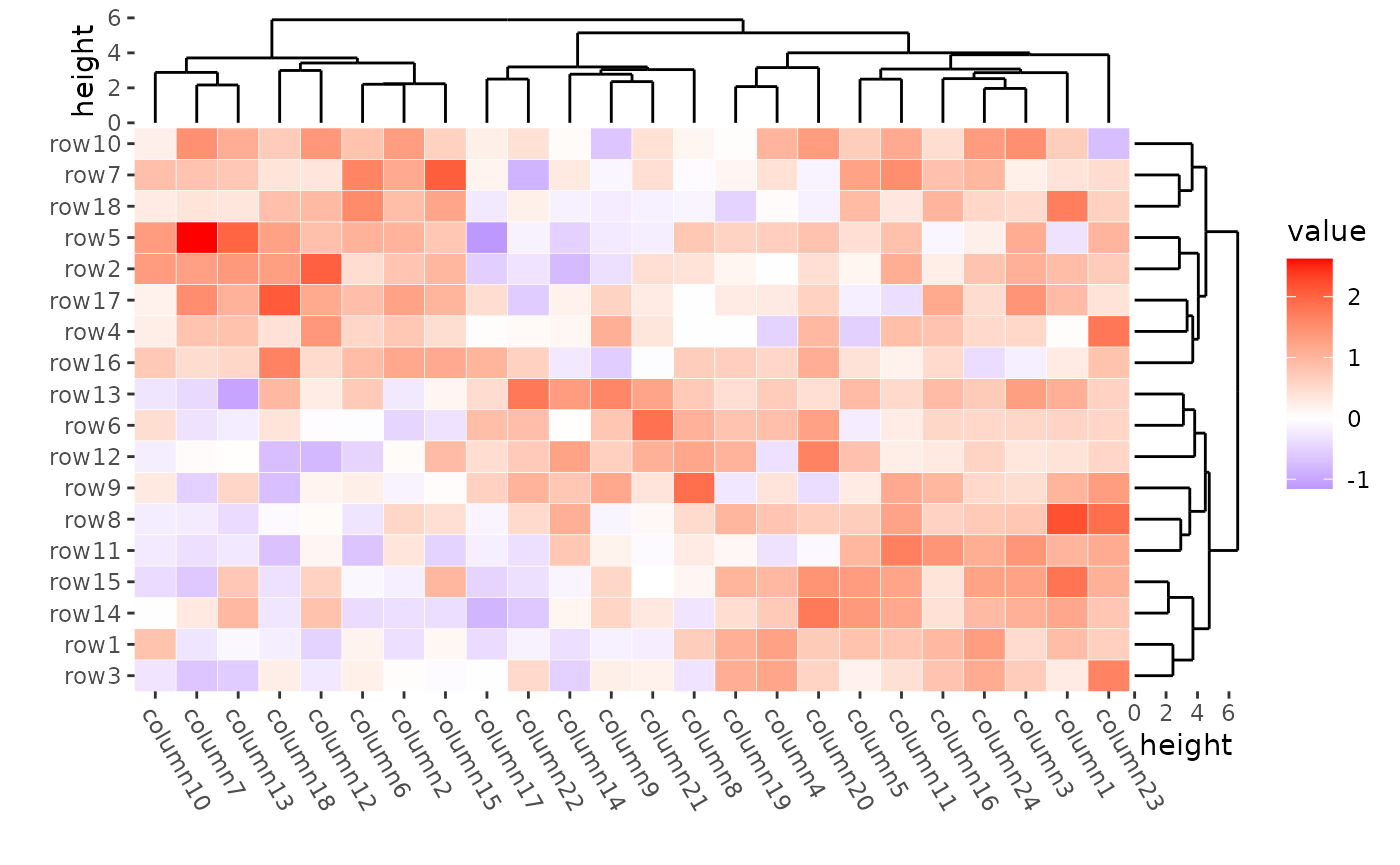
To draw a blank heatmap body:
ggheatmap(mat, filling = NULL) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) &
theme(plot.margin = margin())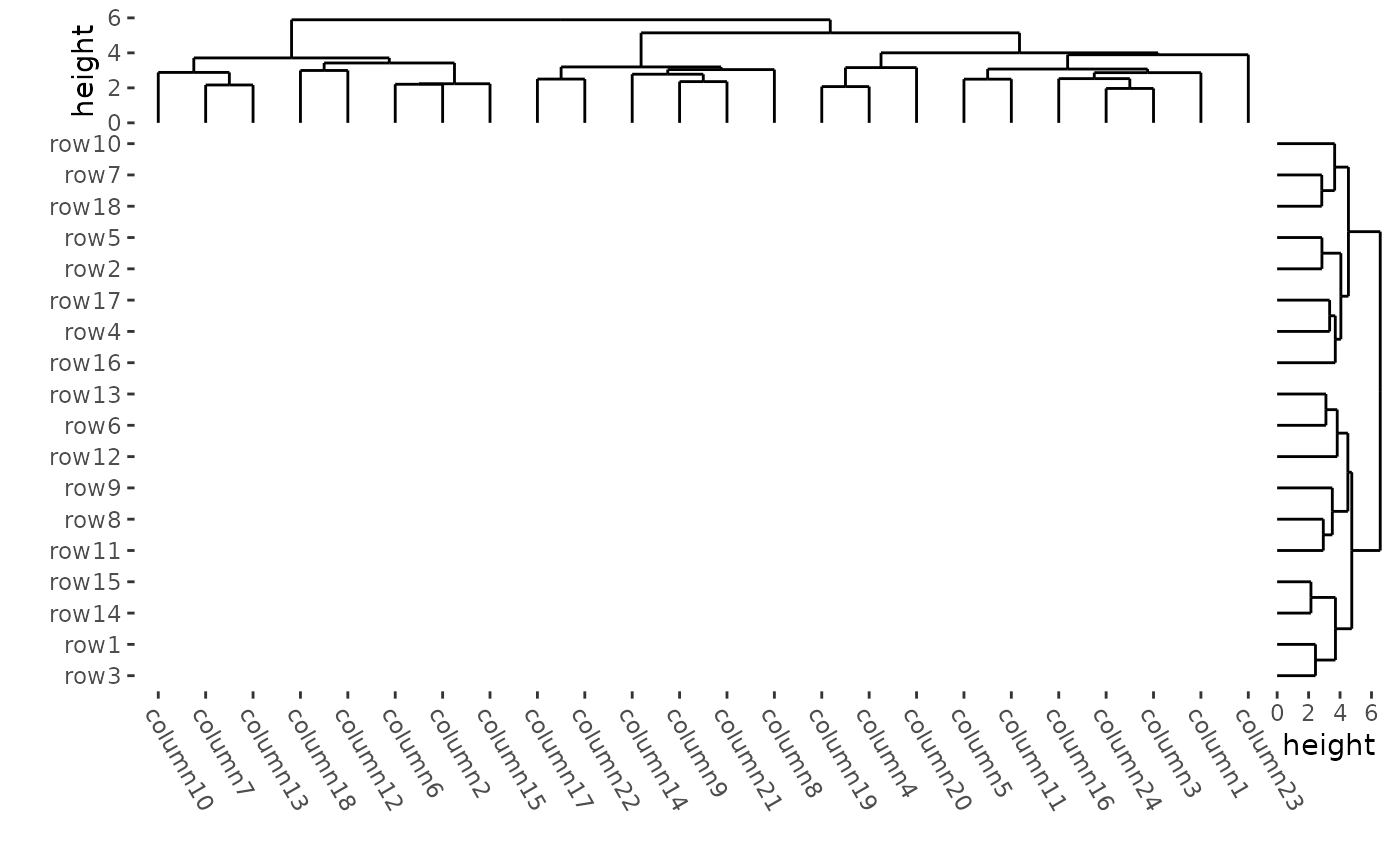
Titles
We can use patch_titles() to add titles around each
border of the plot. You can use theme() to control the text
appearance.
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
patch_titles(right = "I am a row title") +
theme(plot.patch_title.right = element_text(face = "bold", size = 16)) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
patch_titles(top = "I am a column title") +
theme(plot.patch_title.top = element_text(face = "bold", size = 16)) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`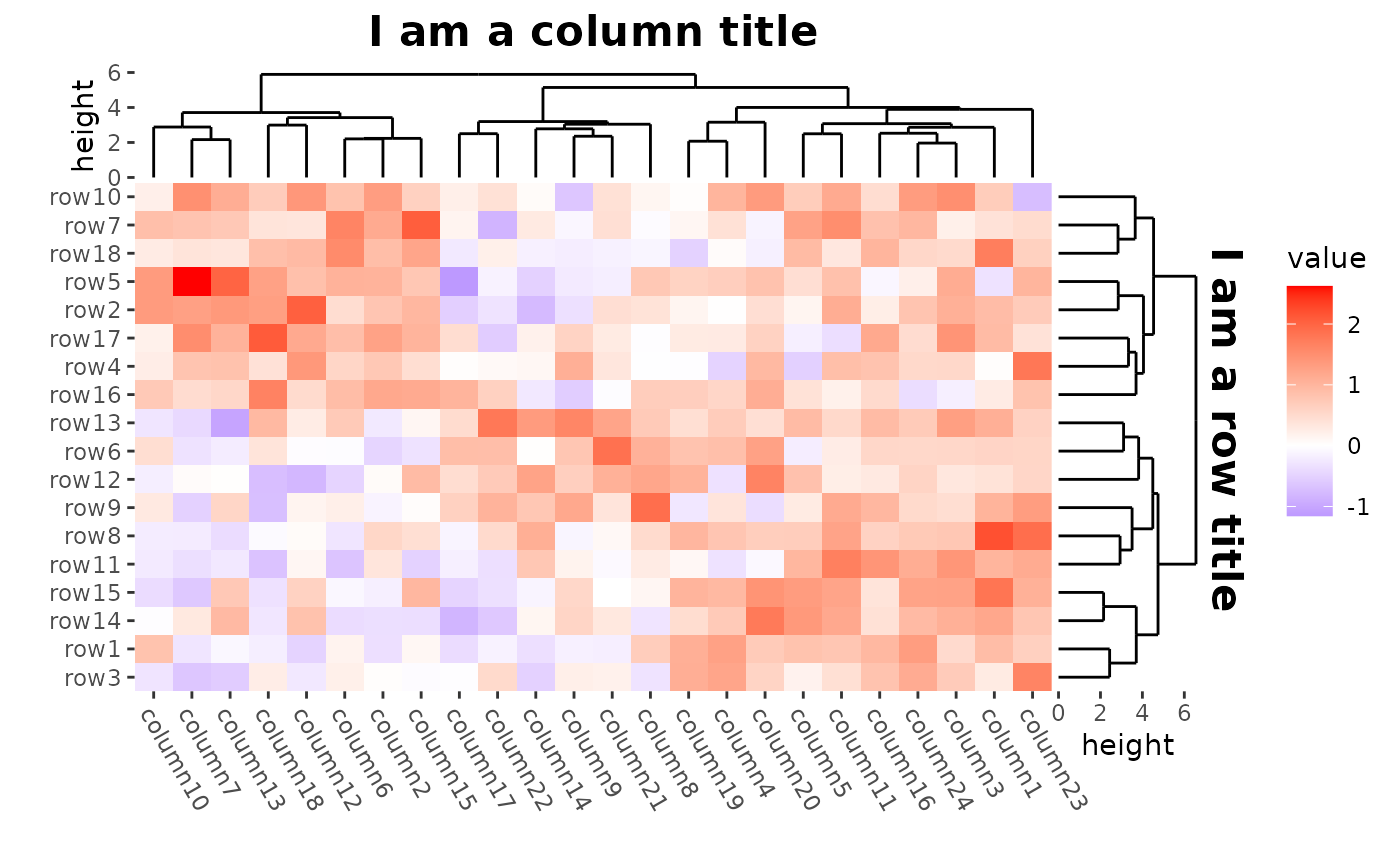
Clustering
Distance methods
# ComplexHeatmap::Heatmap(mat,
# name = "mat", clustering_distance_rows = "pearson",
# column_title = "pre-defined distance method (1 - pearson)"
# )
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_dendro(distance = "pearson", reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
patch_titles(top = "pre-defined distance method (1 - pearson)") +
theme(plot.patch_title.top = element_text(face = "bold", size = 16)) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`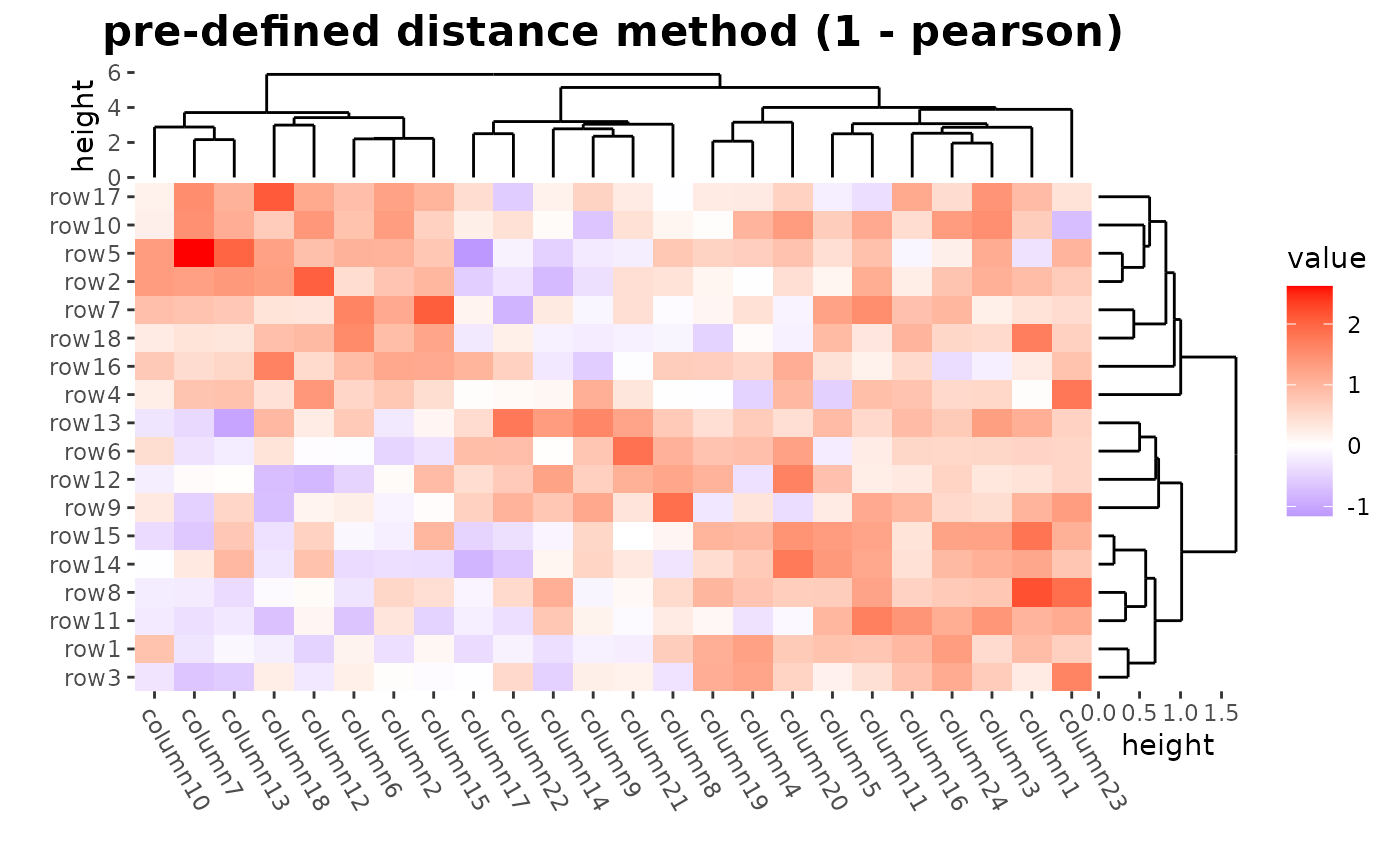
# ComplexHeatmap::Heatmap(mat,
# name = "mat", clustering_distance_rows = function(m) dist(m),
# column_title = "a function that calculates distance matrix"
# )
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_dendro(distance = dist, reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
patch_titles(top = "a function that calculates distance matrix") +
theme(plot.patch_title.top = element_text(face = "bold", size = 16)) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`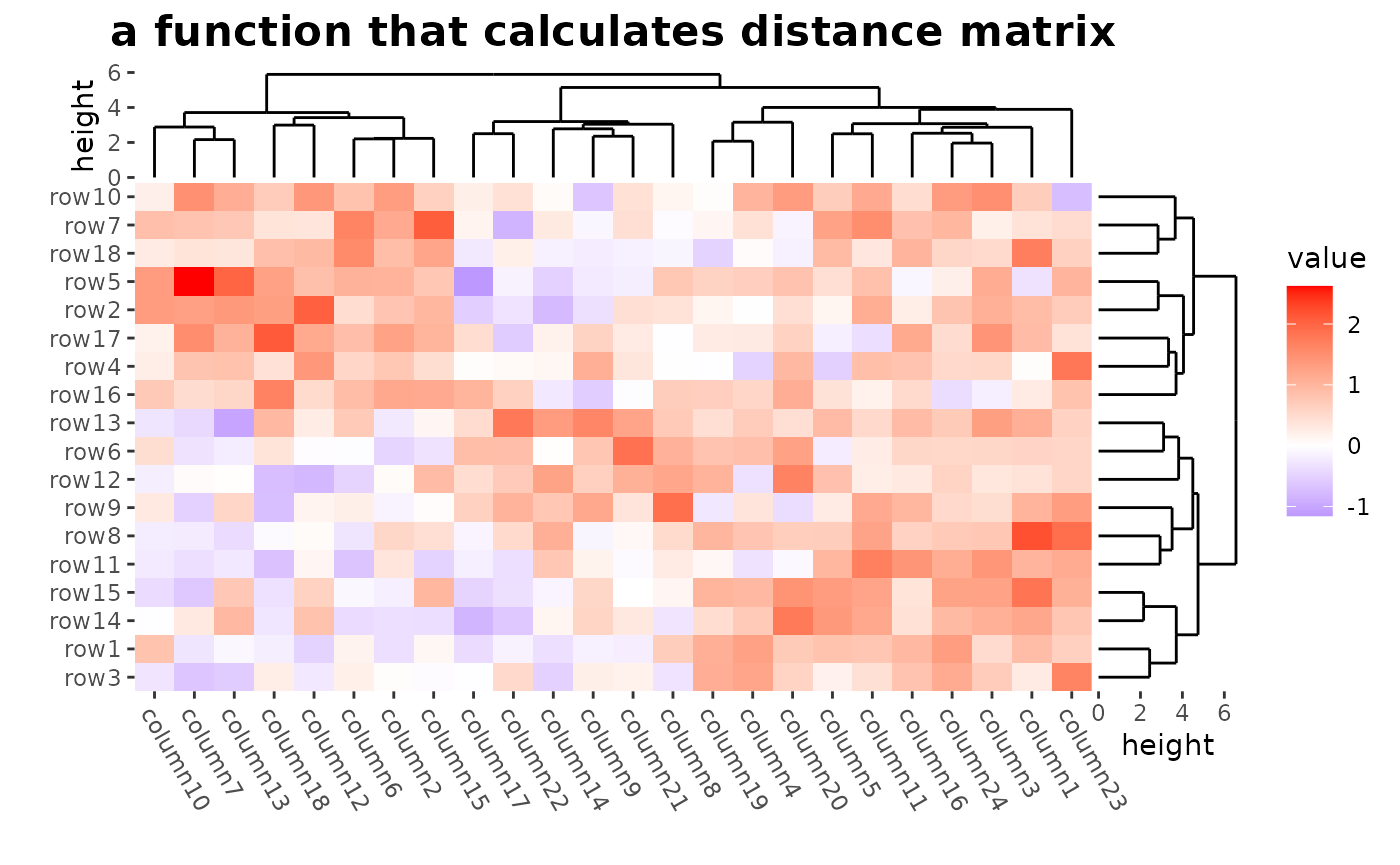
Clustering methods
Method to perform hierarchical clustering can be specified by
method argument, Possible methods are those supported in
hclust() function.
# ComplexHeatmap::Heatmap(mat,
# name = "mat",
# clustering_method_rows = "single"
# )
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_dendro(method = "single", reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`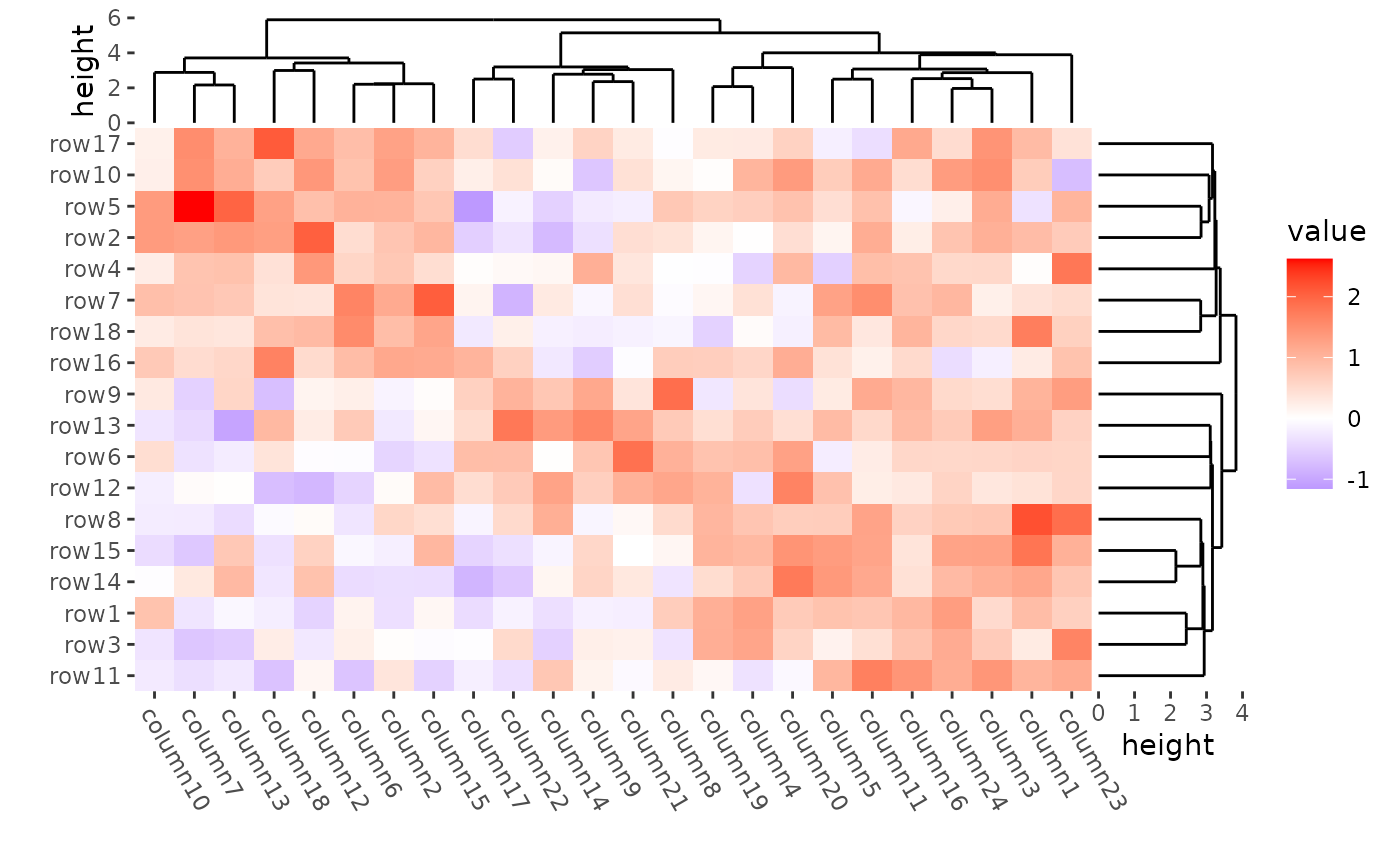
Use distance = NULL if you don’t want to calculate the
distance.
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_dendro(
distance = NULL, method = cluster::diana,
reorder_dendrogram = TRUE
) +
anno_top(size = unit(15, "mm")) +
align_dendro(
distance = NULL, method = cluster::agnes,
reorder_dendrogram = TRUE
) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`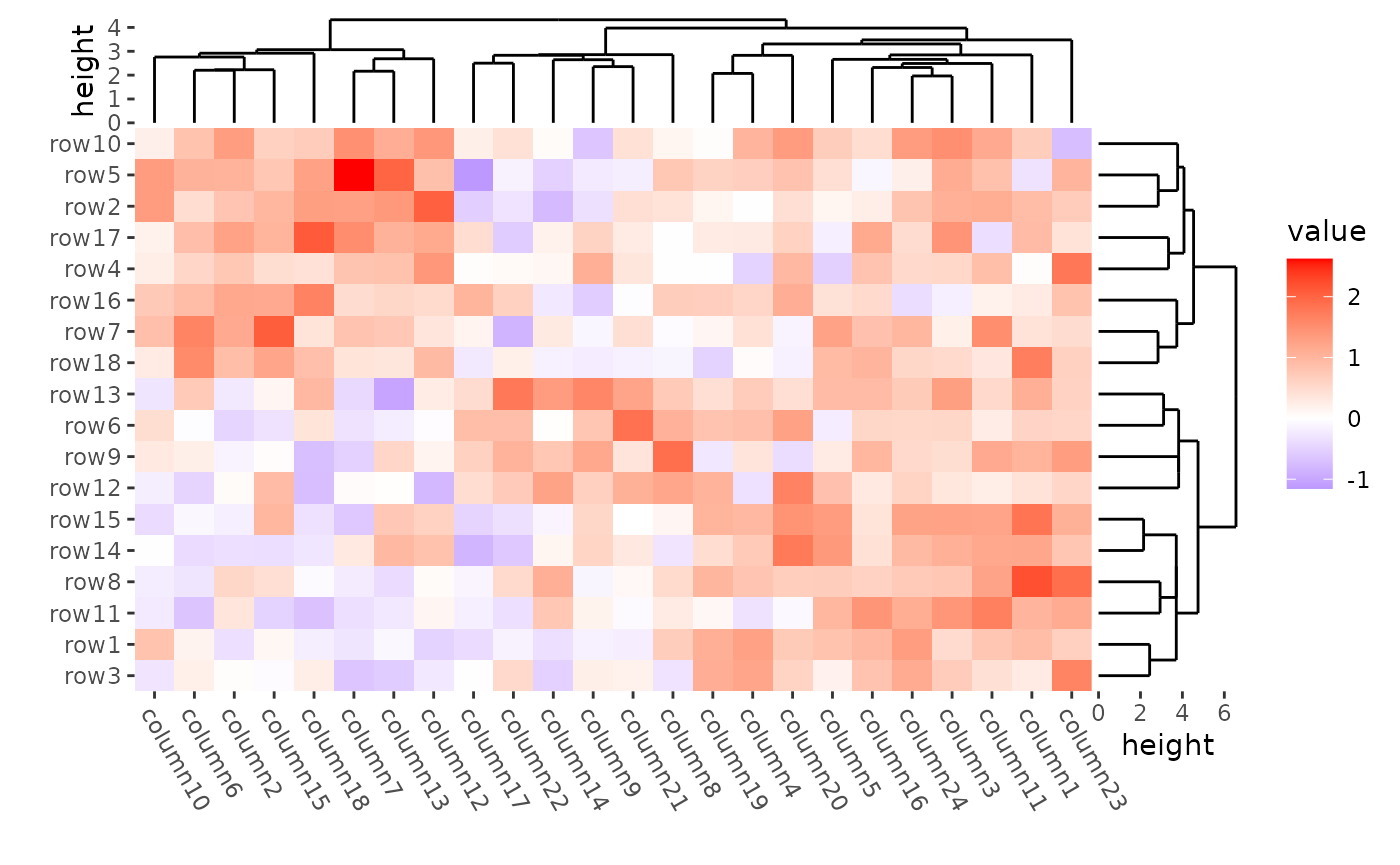
Render dendrograms
It’s easy for ggalign to color the branches by setting
the color mapping, since ggalign will add the
cutree() results into the underlying data.
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_dendro(aes(color = branch), k = 2L, reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`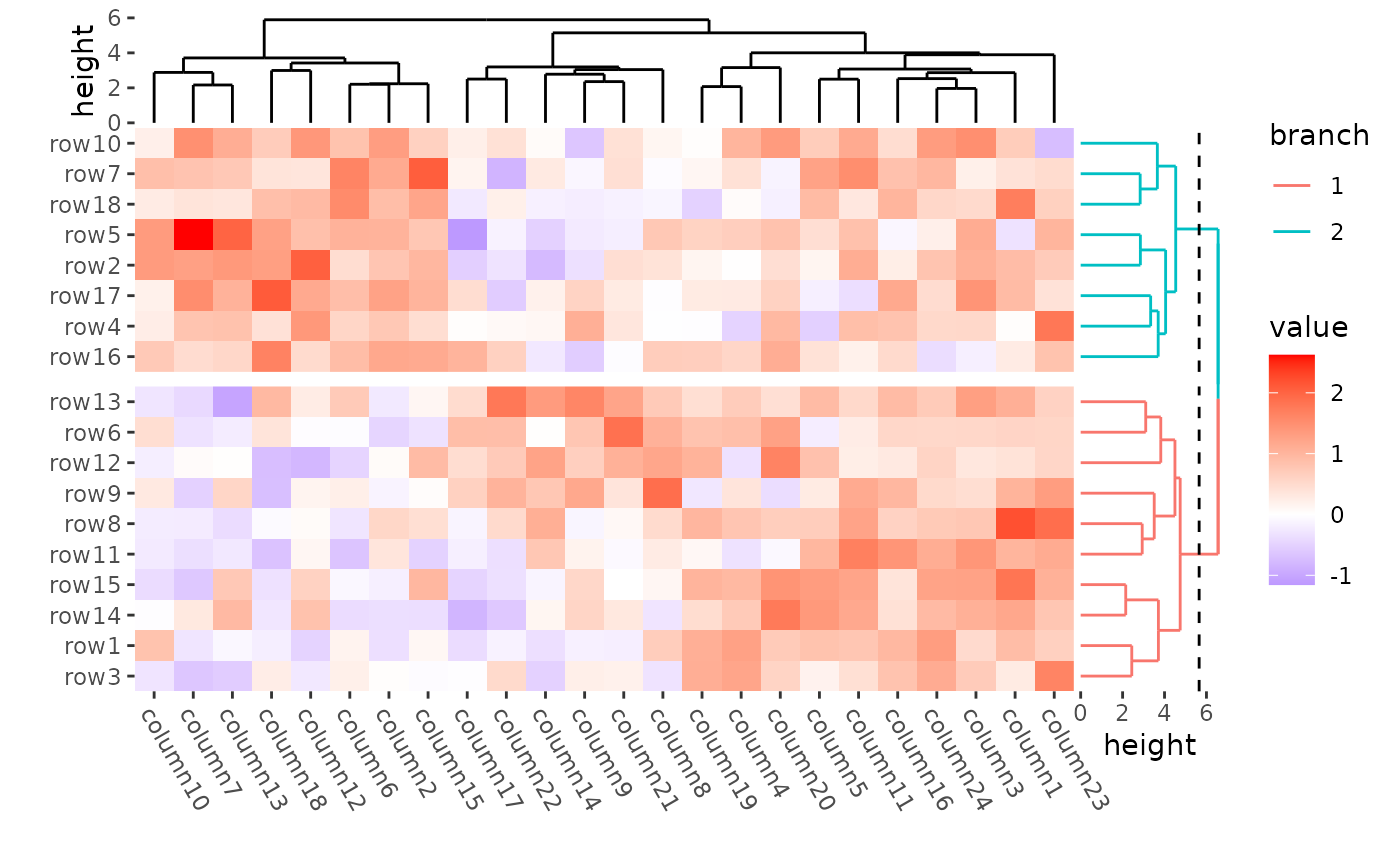
Set row and column orders
We can use align_order() to set the order.
# ComplexHeatmap::Heatmap(mat,
# name = "mat",
# row_order = order(as.numeric(gsub("row", "", rownames(mat)))),
# column_order = order(as.numeric(gsub("column", "", colnames(mat)))),
# column_title = "reorder matrix"
# )
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_order(as.numeric(gsub("row", "", rownames(mat)))) +
anno_top(size = unit(15, "mm")) +
align_order(as.numeric(gsub("column", "", colnames(mat)))) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`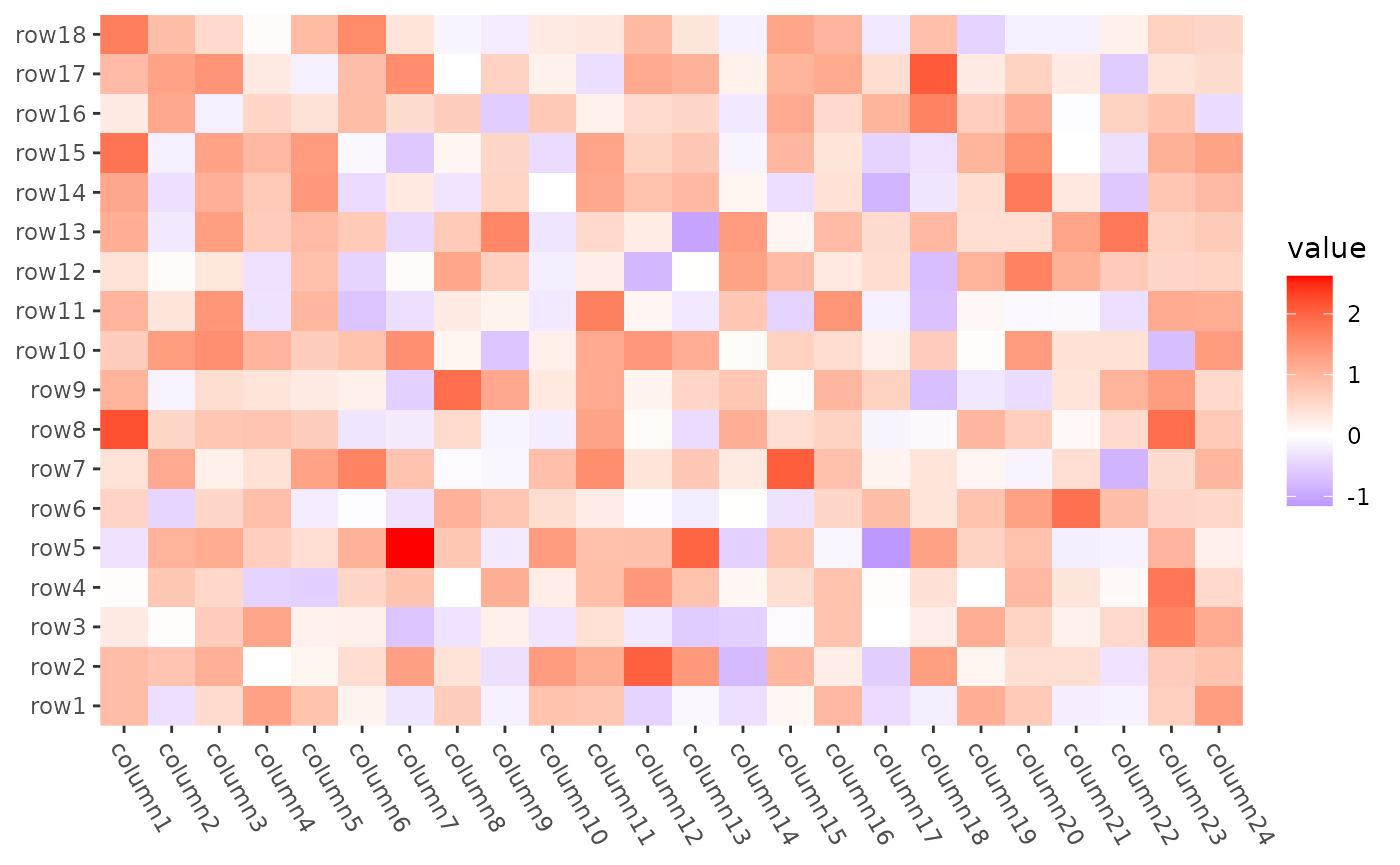
Seriation
align_reorder() can directly take the
seriate() function as the input and extract the ordering
information.
mat2 <- max(mat) - mat
ggheatmap(mat2) +
scale_fill_gradient2(low = "blue", high = "red", midpoint = 2L) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_reorder(seriation::seriate, method = "BEA_TSP") +
anno_top(size = unit(15, "mm")) +
align_reorder(seriation::seriate, method = "BEA_TSP") &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`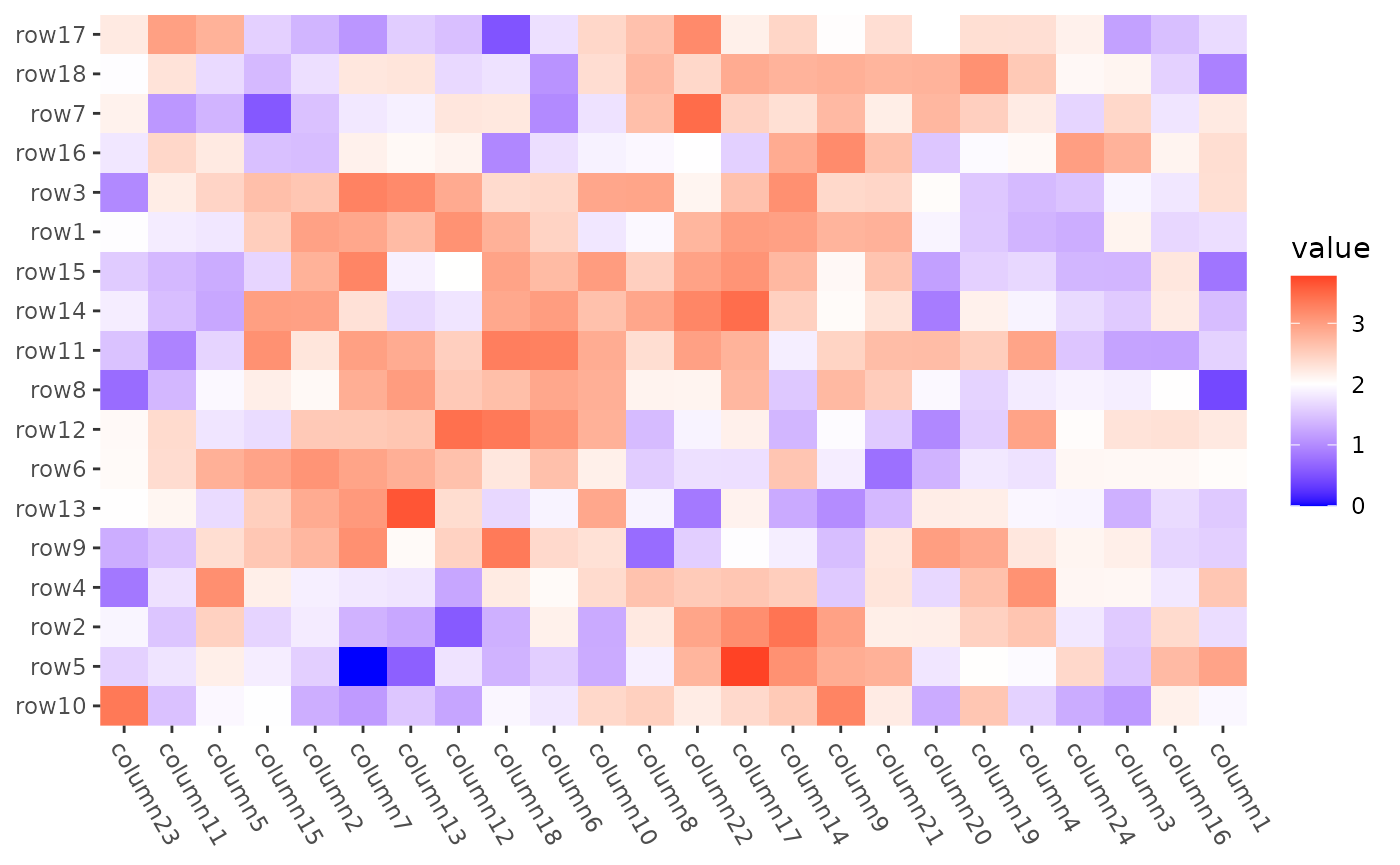
The above code will execute seriate() twice—once for
each dimension. However, since a single run of seriate()
can provide the ordering for both dimensions, we can manually extract
the ordering indices to avoid redundancy.
o <- seriation::seriate(mat2, method = "BEA_TSP")
ggheatmap(mat2) +
scale_fill_gradient2(low = "blue", high = "red", midpoint = 2L) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(20, "mm")) +
align_order(seriation::get_order(o, 1L)) +
anno_top(size = unit(15, "mm")) +
align_order(seriation::get_order(o, 2L)) &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`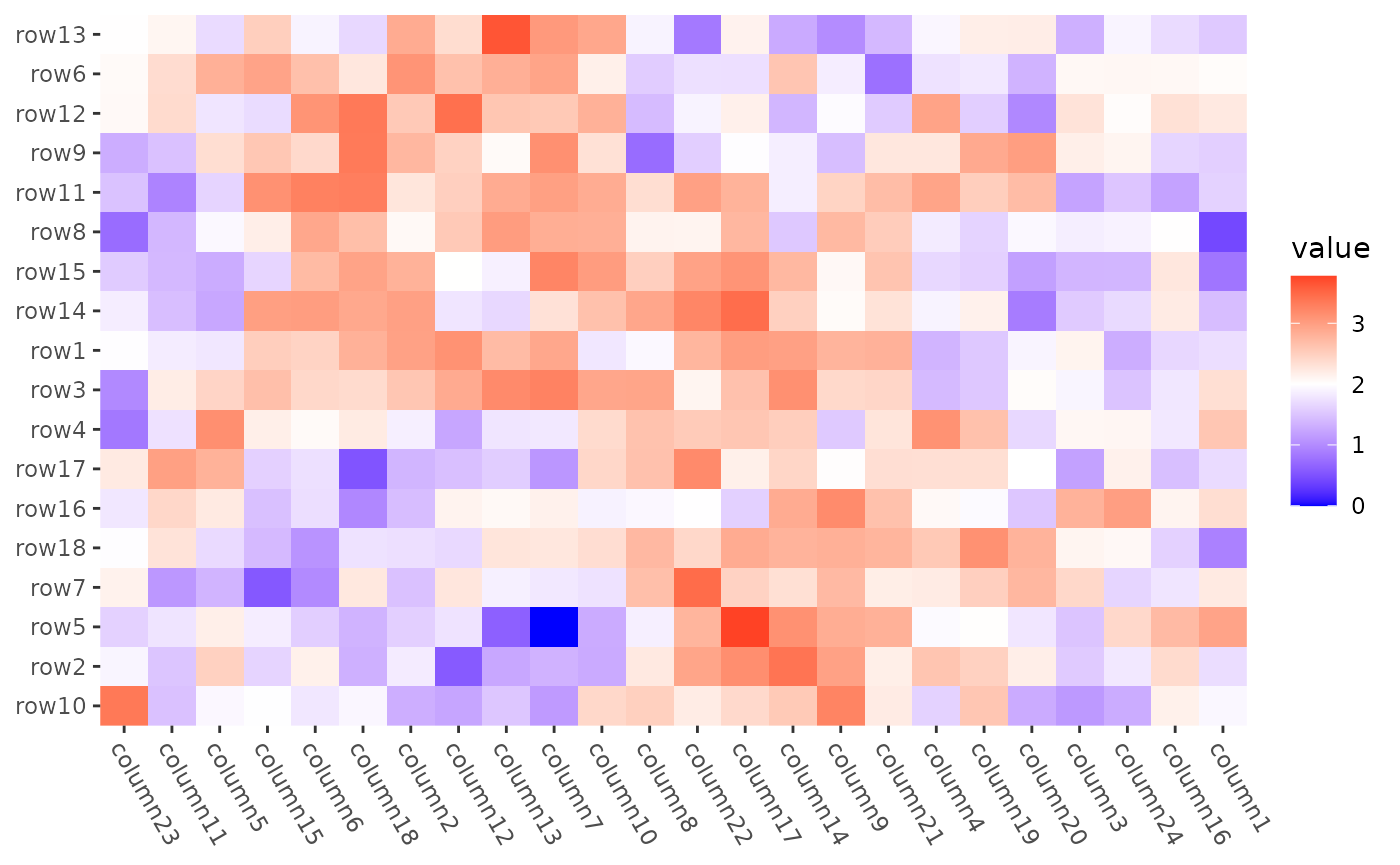 For more use of the
For more use of the seriate() function, please refer to the
seriation
package.
Dimension labels
ggplot2 use scales and theme to control the axis labels,
Please see scales
section for more details.
# ComplexHeatmap::Heatmap(mat,
# name = "mat", row_names_side = "left", row_dend_side = "right",
# column_names_side = "top", column_dend_side = "bottom"
# )
ggheatmap(mat) +
scale_x_continuous(position = "top") +
scale_y_continuous(position = "right") +
theme(axis.text.x = element_text(angle = 60, hjust = 0)) +
anno_left(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
scale_x_continuous(position = "top") +
anno_bottom(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
scale_y_continuous(position = "right") +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`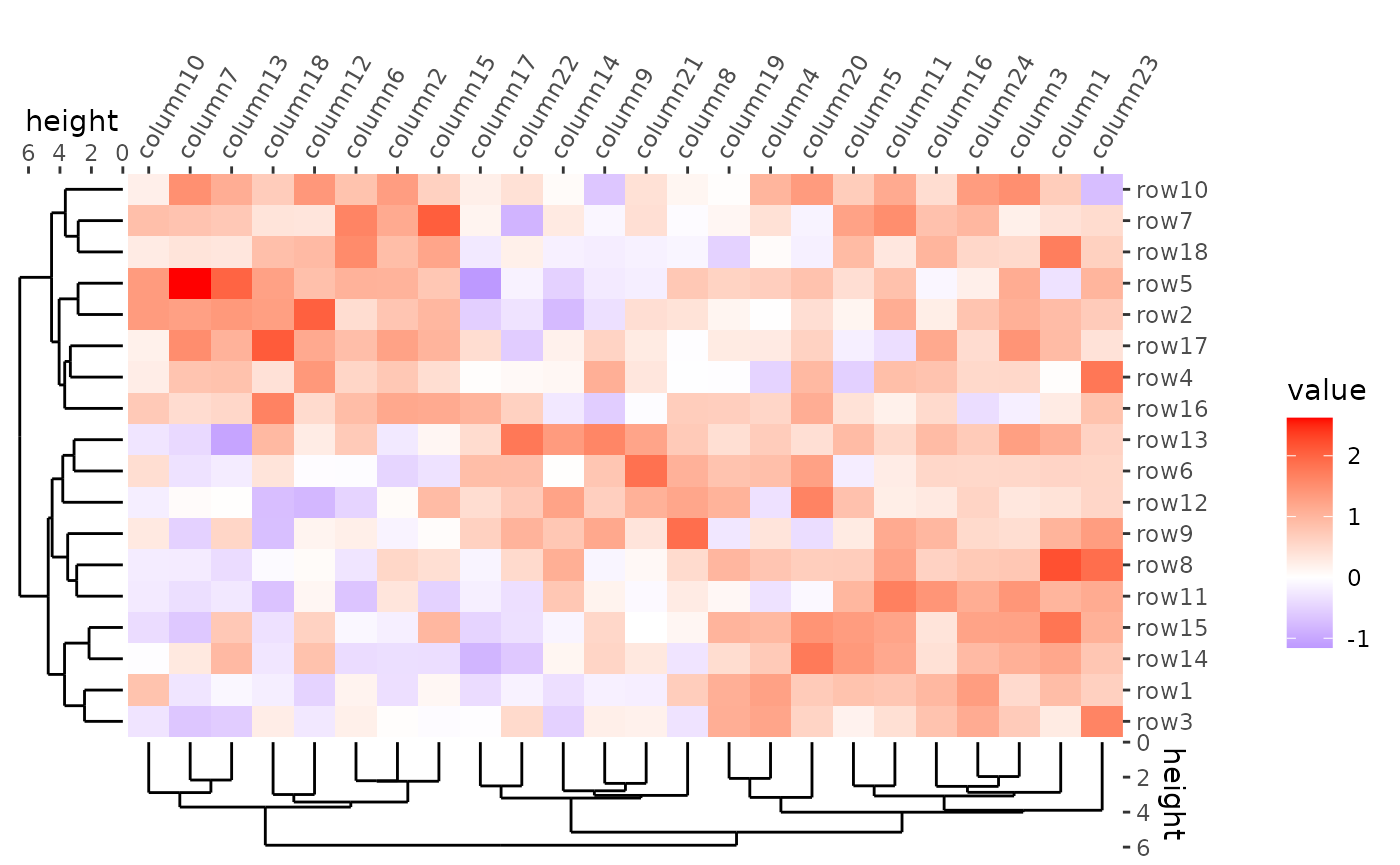
ggheatmap(mat) +
scale_y_continuous(breaks = NULL) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`
ggheatmap(mat) +
theme(
axis.text.x = element_text(angle = -60, hjust = 0),
axis.text.y = element_text(face = "bold", size = 16)
) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`
ggheatmap(mat) +
theme(
axis.text.x = element_text(angle = -60, hjust = 0),
axis.text.y = element_text(
face = "bold", size = 16,
colour = c(rep("red", 10), rep("blue", 8))
)
) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> Warning: Vectorized input to `element_text()` is not officially supported.
#> ℹ Results may be unexpected or may change in future versions of ggplot2.
#> → heatmap built with `geom_tile()`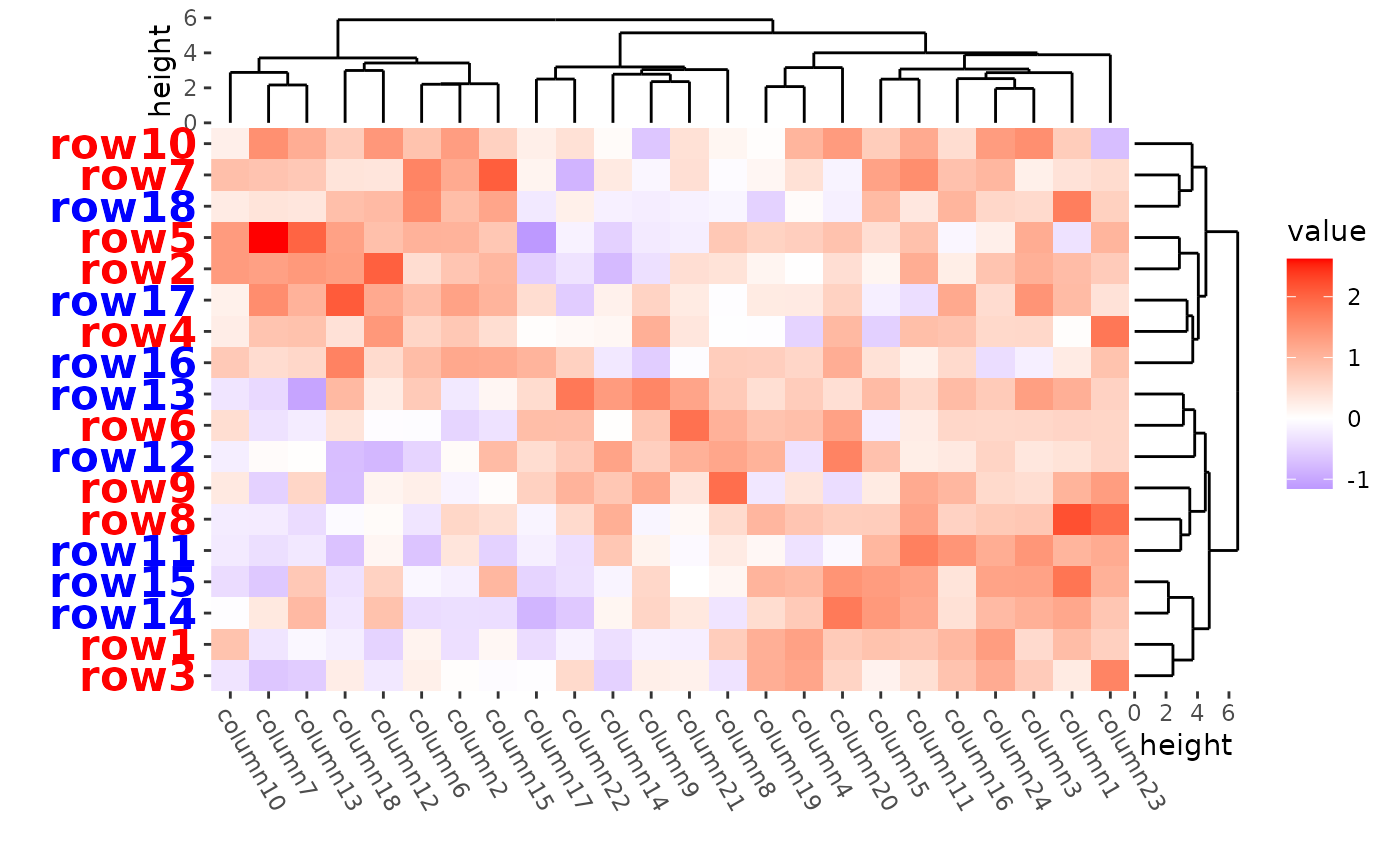
Heatmap split
Split by k-means clustering
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_kmeans(2L) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`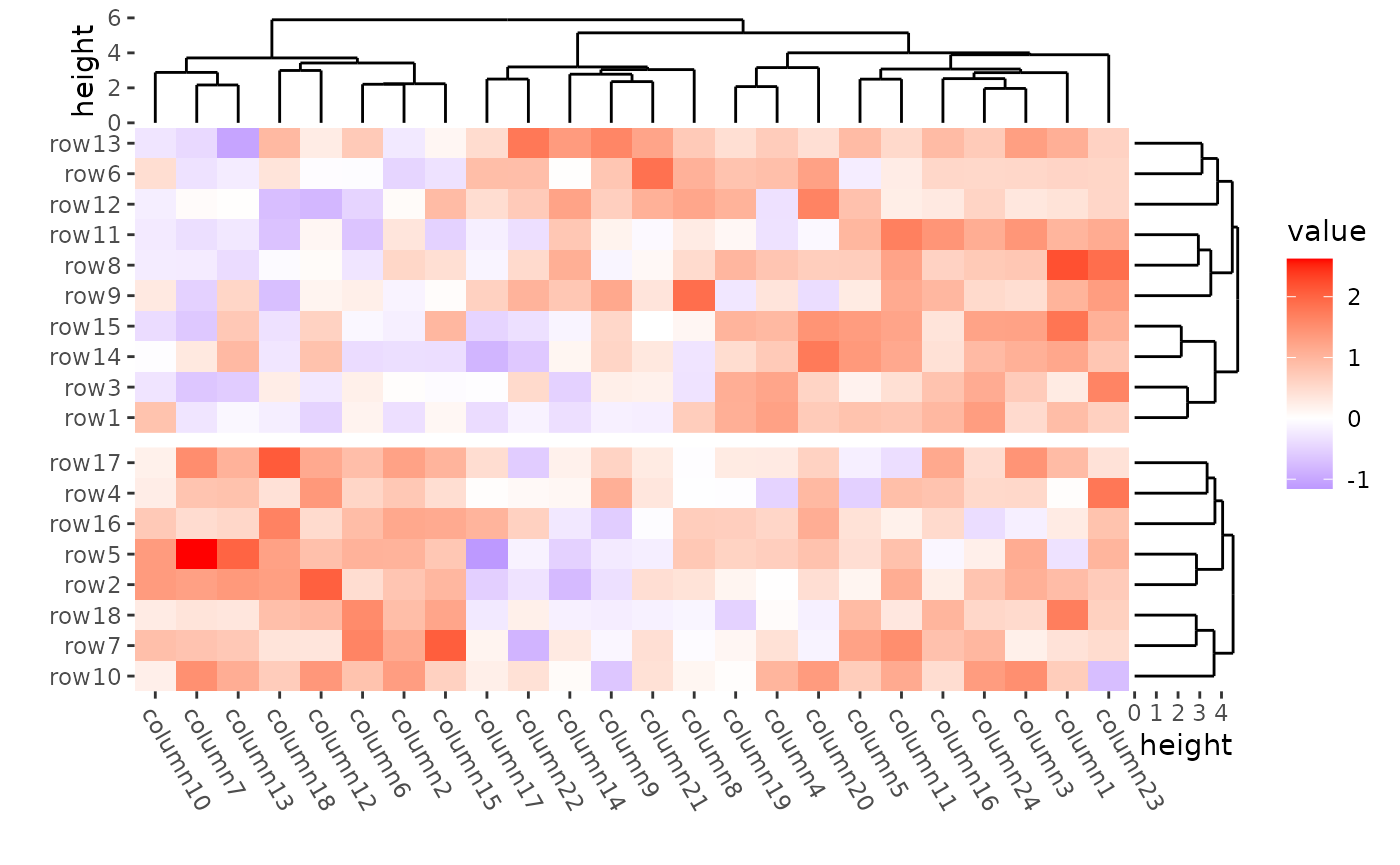
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_kmeans(3L) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`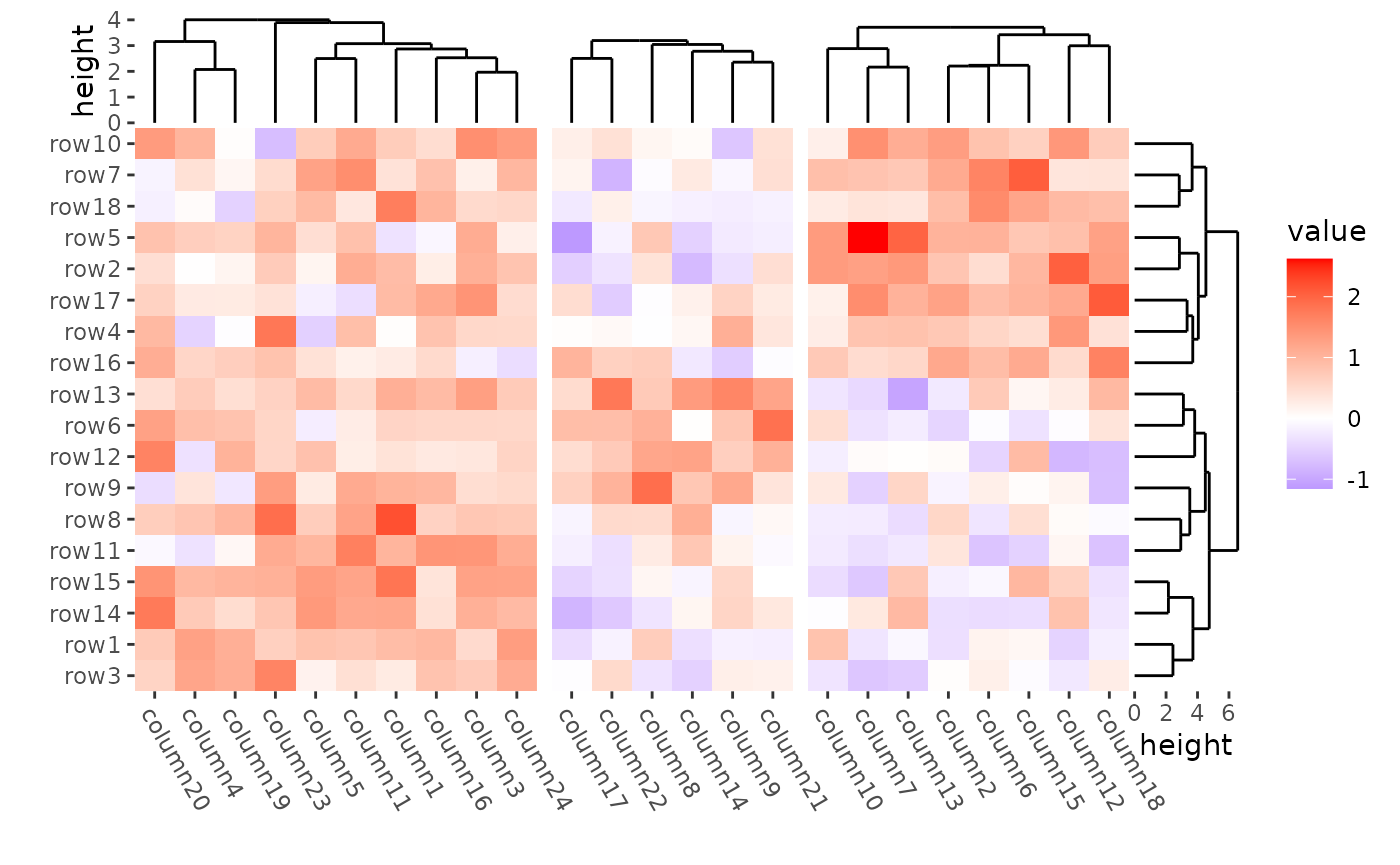
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_kmeans(2L) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_kmeans(3L) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`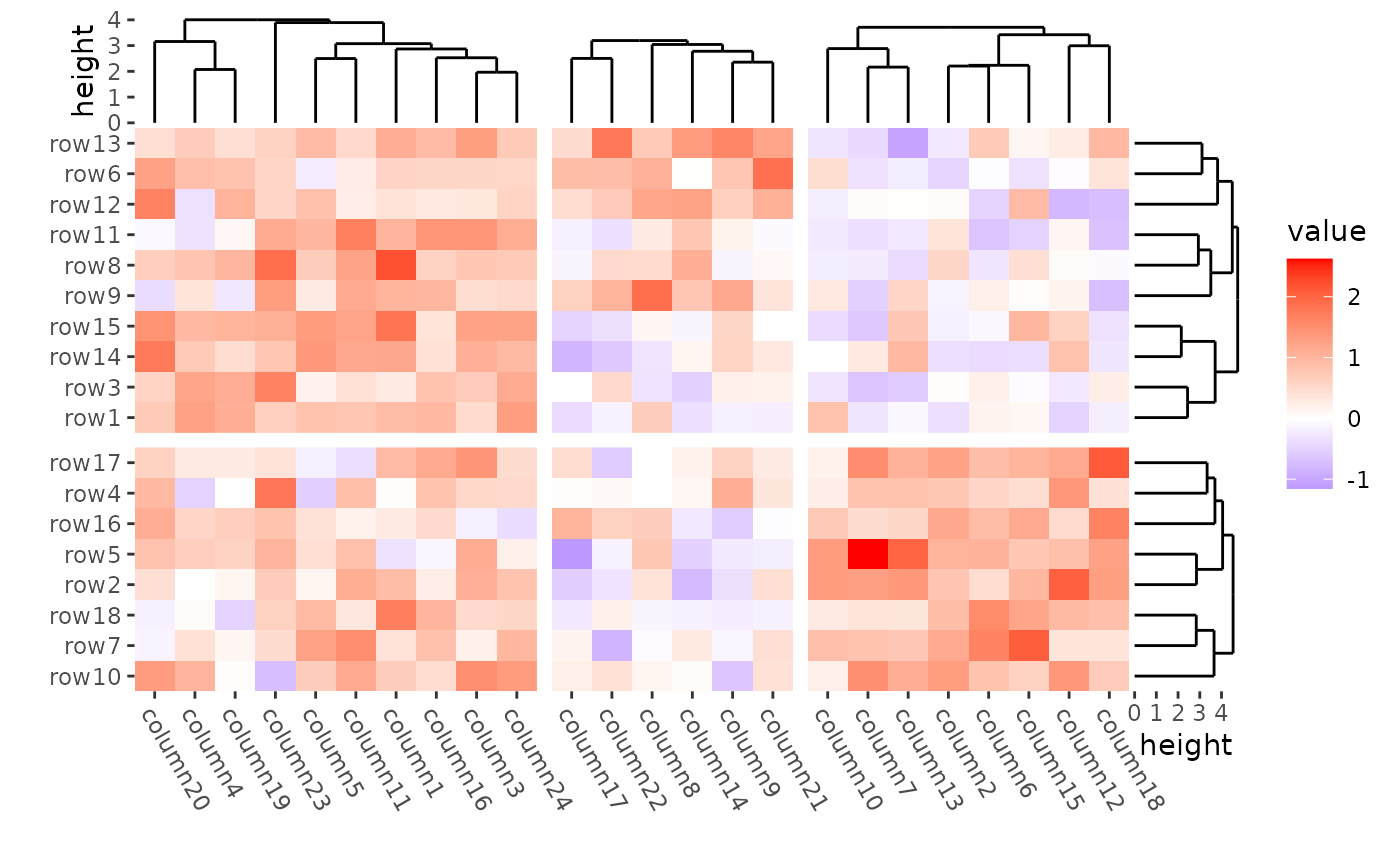
The dendrogram was calculated in each group defined by kmeans.
Split by categorical variables
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_group(rep(c("A", "B"), 9)) +
align_dendro(reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_group(rep(c("C", "D"), 12)) +
align_dendro(reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`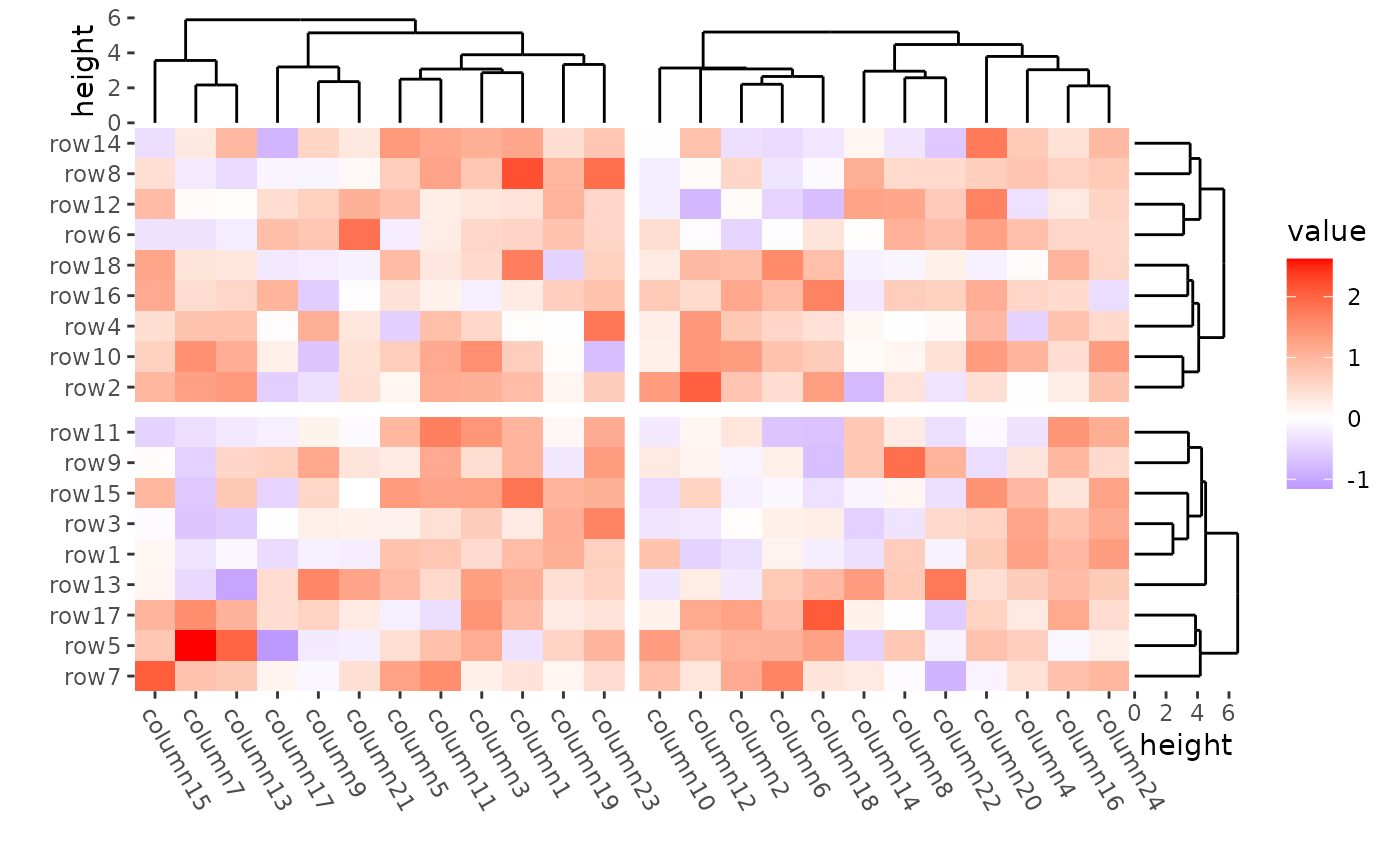
Split by dendrogram
When you splitted by a dendrogram, the cutted height will be indicated with a dashed line.
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(k = 3L, reorder_dendrogram = TRUE) +
anno_top(size = unit(15, "mm")) +
align_dendro(k = 2L, reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`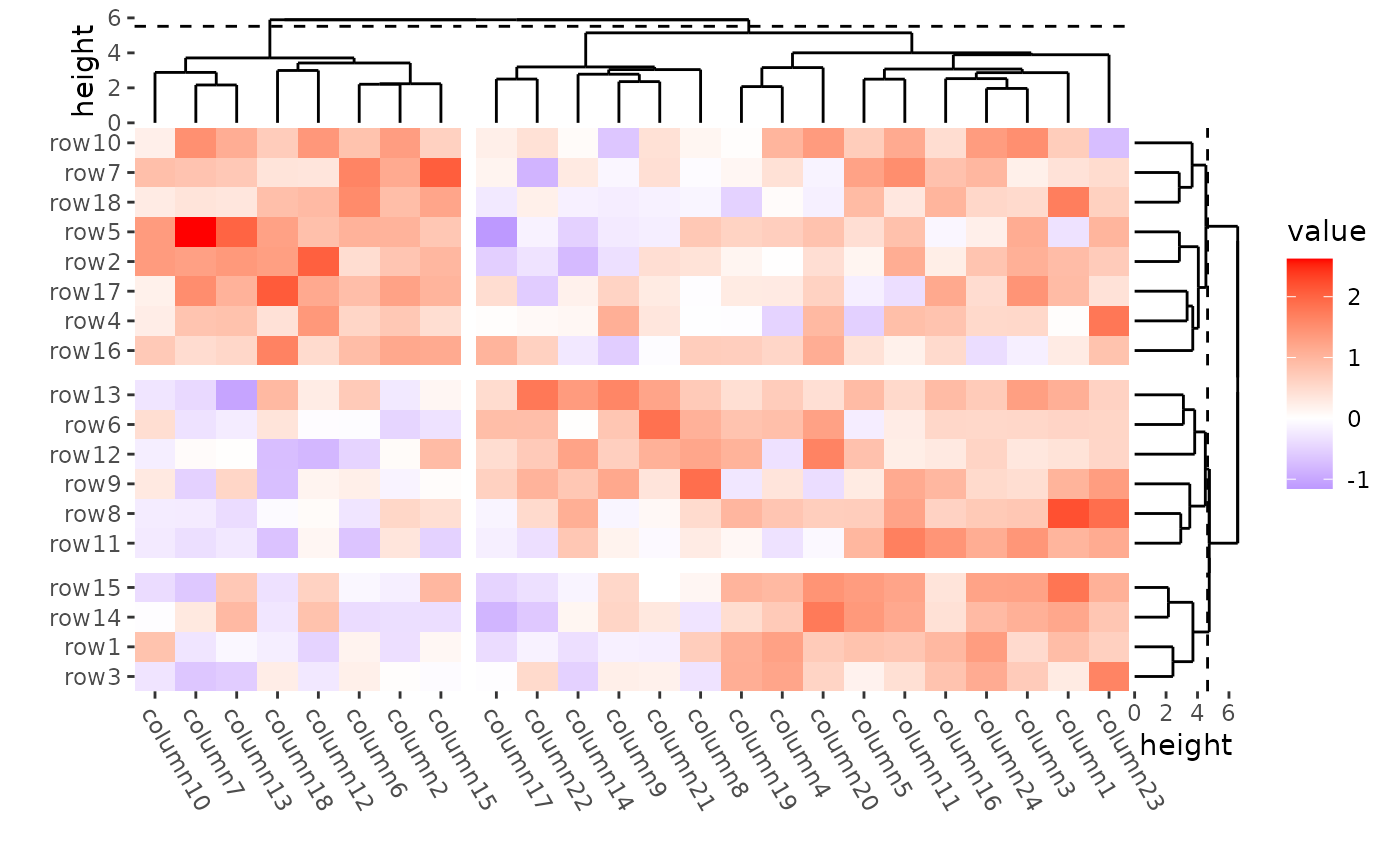
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_dendro(aes(color = branch), k = 3L, reorder_dendrogram = TRUE) +
scale_color_brewer(palette = "Dark2") +
anno_top(size = unit(15, "mm")) +
align_dendro(k = 2L, reorder_dendrogram = TRUE) +
quad_active() &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`
Order of slices (panels)
The order of the panels always follow the factor level. Note: the
merging of dendrogram between ComplexHeatmap and
ggalign is a little different.
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_group(rep(LETTERS[1:3], 6)) +
align_dendro(aes(color = branch),
reorder_dendrogram = TRUE,
reorder_group = TRUE,
merge_dendrogram = TRUE
) +
scale_color_brewer(palette = "Dark2") +
anno_top(size = unit(15, "mm")) +
align_group(rep(letters[1:6], 4)) +
align_dendro(aes(color = branch),
reorder_dendrogram = TRUE,
reorder_group = TRUE,
merge_dendrogram = TRUE
) +
quad_active() -
with_quad(theme(strip.text = element_text()), "tr") &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`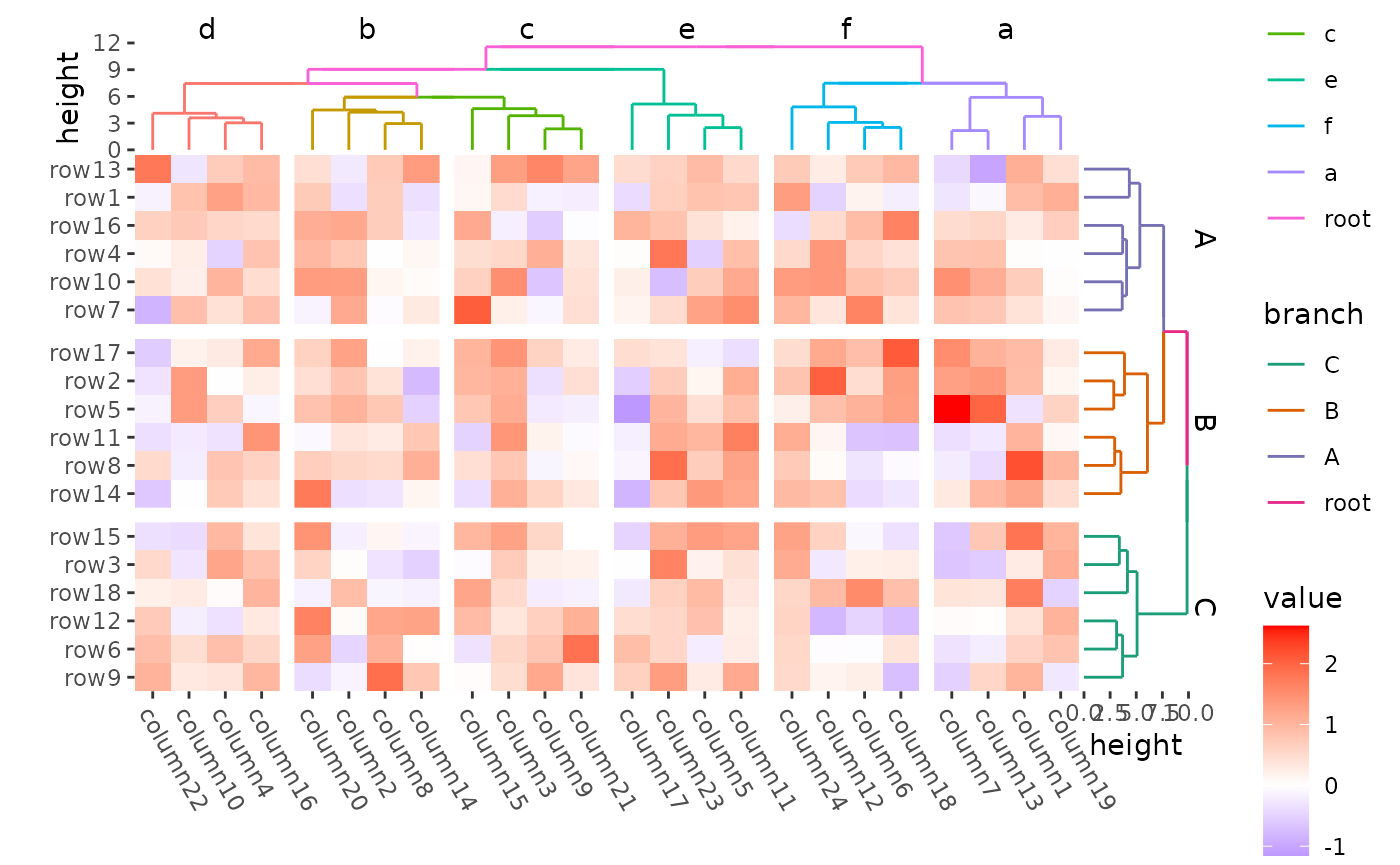
ggheatmap(mat) +
theme(axis.text.x = element_text(angle = -60, hjust = 0)) +
anno_right(size = unit(15, "mm")) +
align_group(rep(LETTERS[1:3], 6)) +
align_dendro(aes(color = branch), reorder_dendrogram = TRUE) +
scale_color_brewer(palette = "Dark2") +
anno_top(size = unit(15, "mm")) +
align_group(rep(letters[1:6], 4)) +
align_dendro(aes(color = branch), reorder_dendrogram = TRUE) +
quad_active() -
with_quad(theme(strip.text = element_text()), "tr") &
theme(plot.margin = margin())
#> → heatmap built with `geom_tile()`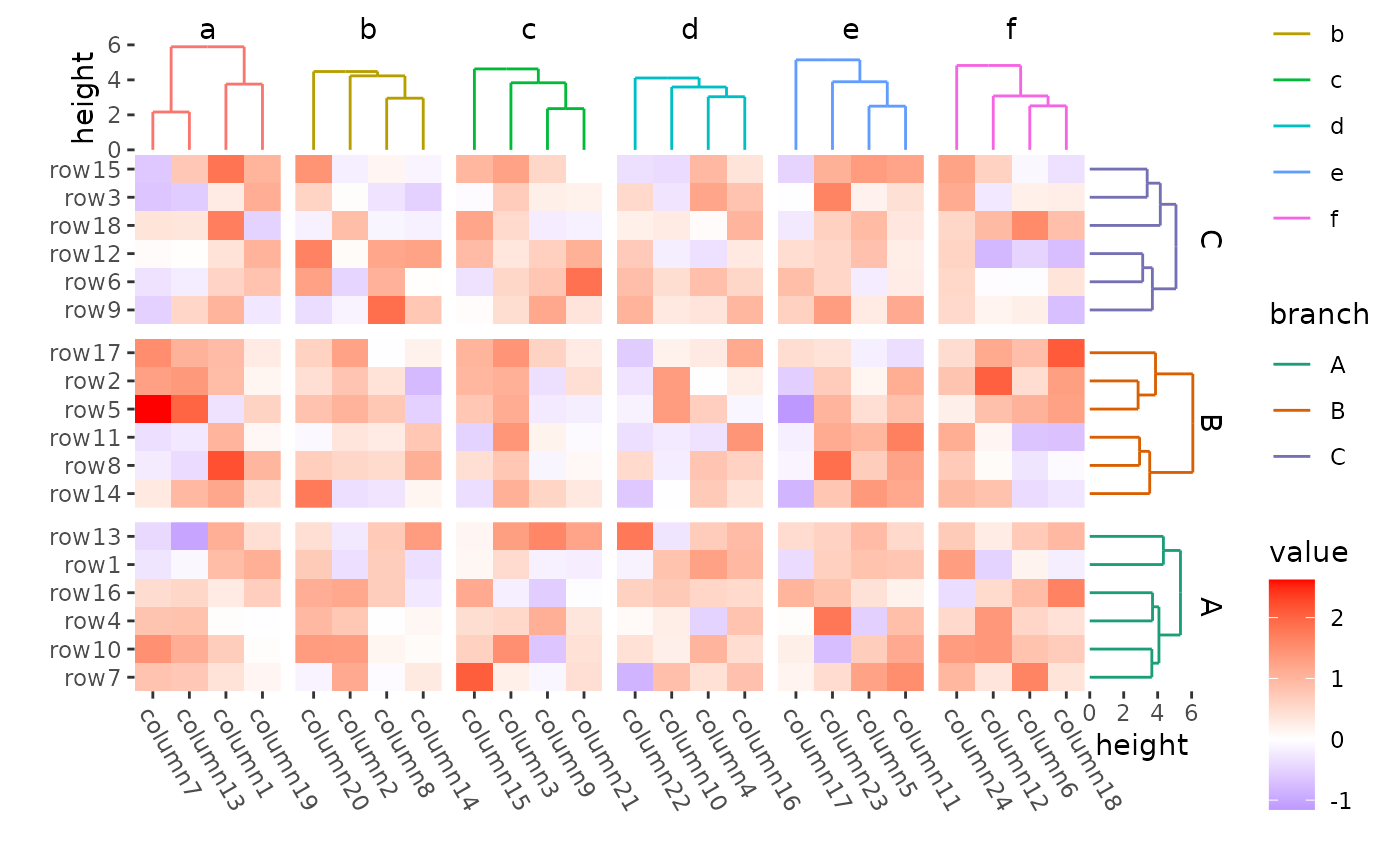
Titles for splitting (facet strip text)
By default, the facet strip text is removed. You can override this
behavior with theme(strip.text = element_text()). Since
align_group() does not create a new plot, the panel title
can only be added to the heatmap plot.
waiting for complete …
Graphic parameters for splitting
ggh4x::facet_grid2(strip = ggh4x::strip_themed(
background_x = list(
element_rect(fill = "red"),
element_rect(fill = "blue"),
element_rect(fill = "green")
)
))
#> <ggproto object: Class FacetGrid2, FacetGrid, Facet, gg>
#> attach_axes: function
#> compute_layout: function
#> draw_back: function
#> draw_front: function
#> draw_labels: function
#> draw_panels: function
#> finish_data: function
#> finish_panels: function
#> init_scales: function
#> map_data: function
#> params: list
#> setup_aspect_ratio: function
#> setup_axes: function
#> setup_data: function
#> setup_panel_table: function
#> setup_params: function
#> shrink: TRUE
#> strip: <ggproto object: Class StripElemental, Strip, gg>
#> assemble_strip: function
#> build_strip: function
#> clip: inherit
#> draw_labels: function
#> elements: list
#> finish_strip: function
#> get_strips: function
#> given_elements: list
#> incorporate_grid: function
#> incorporate_wrap: function
#> init_strip: function
#> params: list
#> setup: function
#> setup_elements: function
#> strips: list
#> super: <ggproto object: Class StripElemental, Strip, gg>
#> train_scales: function
#> vars: function
#> vars_combine: function
#> super: <ggproto object: Class FacetGrid2, FacetGrid, Facet, gg>Session information
sessionInfo()
#> R version 4.4.2 (2024-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 22.04.5 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggalign_0.0.5.9000 ggplot2_3.5.1
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.6 jsonlite_1.8.9 dplyr_1.1.4 compiler_4.4.2
#> [5] tidyselect_1.2.1 cluster_2.1.6 jquerylib_0.1.4 systemfonts_1.1.0
#> [9] scales_1.3.0 textshaping_0.4.1 ggh4x_0.2.8 yaml_2.3.10
#> [13] fastmap_1.2.0 R6_2.5.1 labeling_0.4.3 generics_0.1.3
#> [17] ca_0.71.1 knitr_1.49 iterators_1.0.14 tibble_3.2.1
#> [21] desc_1.4.3 munsell_0.5.1 RColorBrewer_1.1-3 bslib_0.8.0
#> [25] pillar_1.9.0 rlang_1.1.4 utf8_1.2.4 cachem_1.1.0
#> [29] xfun_0.49 fs_1.6.5 sass_0.4.9 TSP_1.2-4
#> [33] registry_0.5-1 cli_3.6.3 withr_3.0.2 pkgdown_2.1.1
#> [37] magrittr_2.0.3 digest_0.6.37 foreach_1.5.2 grid_4.4.2
#> [41] lifecycle_1.0.4 vctrs_0.6.5 evaluate_1.0.1 glue_1.8.0
#> [45] farver_2.1.2 codetools_0.2-20 ragg_1.3.3 fansi_1.0.6
#> [49] colorspace_2.1-1 rmarkdown_2.29 seriation_1.5.7 pkgconfig_2.0.3
#> [53] tools_4.4.2 htmltools_0.5.8.1